Selective Electrochemical Detection of SARS-CoV-2 Using Deep Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. UFC-19 Sensor
2.2. Initial Signature Analysis of SARS-CoV-2 and Comparison with Other Viruses
2.3. Machine Learning and Deep Learning Algorithms
2.3.1. Machine Learning Algorithms with Manual Feature Extraction
2.3.2. Convolution Neural Networks (CNN)
3. Results and Discussion
3.1. Results of Initial Signature Analysis of SARS-CoV-2 and Comparison with Other Viruses
3.2. Machine Learning and Deep Learning Results
3.2.1. Machine Learning
3.2.2. Deep Learning Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Pantelis, G.; Lescure, F.X.; Birgand, G.; Holmes, A.H. Machine learning for clinical decision support in infectious diseases: A narrative review of current applications. Clin. Microbiol. Infect. 2020, 26, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Benjamens, S.; Dhunnoo, P.; Mesko, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef]
- Gambhir, S.; Malik, S.K.; Kumar, Y. The Diagnosis of Dengue Disease: An Evaluation of Three Machine Learning Approaches. Int. J. Healthc. Inf. Syst. Inform. 2018, 13, 19. [Google Scholar] [CrossRef]
- Khan, S.; Ullah, R.; Khan, A.; Ashraf, R.; Ali, H.; Bilal, M.; Saleem, M. Analysis of hepatitis B virus infection in blood sera using Raman spectroscopy and machine learning. Photodiagnosis Photodyn. Ther. 2018, 23, 89–93. [Google Scholar] [CrossRef]
- Jeong, Y.S.; Jeon, M.; Park, J.H.; Kim, M.C.; Lee, E.; Park, S.Y.; Lee, Y.M.; Choi, S.; Park, S.Y.; Park, K.H.; et al. Machine-Learning-Based Approach to Differential Diagnosis in Tuberculous and Viral Meningitis. Infect. Chemother. 2021, 53, 53–62. [Google Scholar] [CrossRef]
- Rohaim, M.A.; Clayton, E.; Sahin, I.; Vilela, J.; Khalifa, M.E.; Al-Natour, M.Q.; Bayoumi, M.; Poirier, A.C.; Branavan, M.; Tharmakulasingam, M.; et al. Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses 2020, 12, 972. [Google Scholar] [CrossRef]
- Benameur, N.; Mahmoudi, R.; Zaid, S.; Arous, Y.; Hmida, B.; Bedoui, M.H. SARS-CoV-2 diagnosis using medical imaging techniques and artificial intelligence: A review. Clin. Imaging 2021, 76, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Lalmuanawma, S.; Hussain, J.; Chhakchhuak, L. Applications of machine learning and artificial intelligence for COVID-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 2020, 139, 110059. [Google Scholar] [CrossRef] [PubMed]
- Elaziz, M.A.; Hosny, K.M.; Salah, A.; Darwish, M.M.; Lu, S.F.; Sahlol, A.T. New machine learning method for image-based diagnosis of COVID-19. PLoS ONE 2020, 15, e0235187. [Google Scholar] [CrossRef] [PubMed]
- Kassani, S.H.; Kassani, P.H.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: A machine learning based approach. Biocybern. Biomed. Eng. 2021, 41, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Razavi-Far, R.; Hallaji, E.; Farajzadeh-Zanjani, M.; Saif, M.; Kia, S.H.; Henao, H.; Capolino, G.A. Information Fusion and Semi-Supervised Deep Learning Scheme for Diagnosing Gear Faults in Induction Machine Systems. IEEE Trans. Ind. Electron. 2019, 66, 6331–6342. [Google Scholar] [CrossRef]
- Kukar, M.; Guncar, G.; Vovko, T.; Podnar, S.; Cernelc, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moskon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef] [PubMed]
- Arpaci, I.; Huang, S.G.; Al-Emran, M.; Al-Kabi, M.N.; Peng, M.F. Predicting the COVID-19 infection with fourteen clinical features using machine learning classification algorithms. Multimed. Tools Appl. 2021, 80, 11943–11957. [Google Scholar] [CrossRef]
- Ma, S.M.; Lai, X.Q.; Chen, Z.; Tu, S.H.; Qin, K. Clinical characteristics of critically ill patients co-infected with SARS-CoV-2 and the influenza virus in Wuhan, China. Int. J. Infect. Dis. 2020, 96, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Colizzi, M.; Bortoletto, R.; Silvestri, M.; Mondini, F.; Puttini, E.; Cainelli, C.; Gaudino, R.; Ruggeri, M.; Zoccante, L. Medically unexplained symptoms in the times of COVID-19 pandemic: A case-report. Brain Behav. Immun.-Health 2020, 5, 100073. [Google Scholar] [CrossRef]
- Dai, W.C.; Zhang, H.W.; Yu, J.; Xu, H.J.; Chen, H.; Luo, S.P.; Zhang, H.; Liang, L.H.; Wu, X.L.; Lei, Y.; et al. CT Imaging and Differential Diagnosis of COVID-19. Can. Assoc. Radiol. J.-J. L Assoc. Can. Radiol. 2020, 71, 195–200. [Google Scholar] [CrossRef]
- Ramanujam, A.; Almodovar, S.; Botte, G.G. Ultra-Fast Electrochemical Sensor for Point-of-Care COVID-19 Diagnosis Using Non-Invasive Saliva Sampling. Processes 2021, 9, 1236. [Google Scholar] [CrossRef]
- Lu, F.; Gecgel, O.; Ramanujam, A.; Botte, G.G. SARS-CoV-2 Surveillance in Indoor Air Using Electrochemical Sensor for Continuous Monitoring and Real-Time Alerts. Biosensors 2022, 12, 523. [Google Scholar] [CrossRef]
- Xu, J.B.; Zhao, S.Z.; Teng, T.S.; Abdalla, A.E.; Zhu, W.; Xie, L.X.; Wang, Y.L.; Guo, X.Q. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 2020, 12, 224. [Google Scholar] [CrossRef]
- Yousefi, H.; Mahmud, A.; Chang, D.R.; Das, J.; Gomis, S.; Chen, J.B.; Wang, H.S.; Been, T.; Yip, L.; Coomes, E.; et al. Detection of SARS-CoV-2 Viral Particles Using Direct, Reagent-Free Electrochemical Sensing. J. Am. Chem. Soc. 2021, 143, 1722–1727. [Google Scholar] [CrossRef]
- Kim, H.Y.; Lee, J.H.; Kim, M.J.; Park, S.C.; Choi, M.; Lee, W.; Ku, K.B.; Kim, B.T.; Park, E.C.; Kim, H.G.; et al. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2021, 175, 112868. [Google Scholar] [CrossRef] [PubMed]
- Botte, G.G.; Ramanujam, A. Rapid Viral Diagnostic Sensor. US11060995B1, 13 July 2021. [Google Scholar]
- US-FDA. Emergency Use Authorization (EUA) Summary COVID-19 RT-PCR Test. Ph.D. Thesis, Laboratory Corporation of America, Burlington, NC, USA, 2021.
- ATCC Heat-Inactivated SARS-CoV-2 (ATCC® VR-1986HK™). Available online: https://www.atcc.org/en/Global/Products/VR-1986HK.aspx#characteristics (accessed on 23 May 2021).
- ZeptoMetrix NATtrol™ Coronavirus SARS Stock (Qualitative) (1 mL) CATALOG# NATSARS-ST. Available online: https://www.zeptometrix.com/products/nattrol-coronavirus-sars-stock-qualitative-1-ml (accessed on 11 October 2021).
- ZeptoMetrix Coronavirus (Strain: OC43) Culture Fluid (Heat Inactivated) (1 mL) CATALOG# 0810024CFHI. Available online: https://www.zeptometrix.com/products/coronavirus-strain-oc43-culture-fluid-heat-inactivated-1-ml (accessed on 25 October 2021).
- ZeptoMetrix MERS-CoV Stock (Qualitative) (1 mL) CATALOG# NATMERS-ST. Available online: https://www.zeptometrix.com/products/nattrol-mers-cov-stock-qualitative-1-ml (accessed on 25 October 2021).
- ZeptoMetrix Influenza A H1N1pdm (NY/02/09) Culture Fluid (Heat Inactivated) (1 mL) CATALOG# 0810109CFNHI. Available online: https://www.zeptometrix.com/products/influenza-a-h1n1pdm-ny-02-09-culture-fluid-heat-inactivated-1-ml (accessed on 25 October 2021).
- Bharti, R.; Khamparia, A.; Shabaz, M.; Dhiman, G.; Pande, S.; Singh, P. Prediction of Heart Disease Using a Combination of Machine Learning and Deep Learning. Comput. Intell. Neurosci. 2021, 2021, 8387680. [Google Scholar] [CrossRef] [PubMed]
- Gecgel, O.; Ekwaro-Osire, S.; Dias, J.P.; Serwadda, A.; Alemayehu, F.M.; Nispel, A.; IEEE. Gearbox Fault Diagnostics Using Deep Learning with Simulated Data. In Proceedings of the IEEE International Conference on Prognostics and Health Management (ICPHM), San Francisco, CA, USA, 17–20 June 2019. [Google Scholar]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Scikit-Learn Machine Learning in Python. Available online: https://scikit-learn.org/stable/index.html (accessed on 21 October 2021).
- Python. Available online: https://www.python.org/ (accessed on 12 December 2020).
- Pennacchio, A.; Giampaolo, F.; Piccialli, F.; Cuomo, S.; Notomista, E.; Spinelli, M.; Amoresano, A.; Piscitelli, A.; Giardina, P. A machine learning-enhanced biosensor for mercury detection based on an hydrophobin chimera. Biosens. Bioelectron. 2022, 196, 113696. [Google Scholar] [CrossRef] [PubMed]
- Otchere, D.A.; Ganat, T.O.A.; Ojero, J.O.; Tackie-Otoo, B.N.; Taki, M.Y. Application of gradient boosting regression model for the evaluation of feature selection techniques in improving reservoir characterisation predictions. J. Pet. Sci. Eng. 2022, 208, 109244. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. Elife 2020, 9, e57309. [Google Scholar] [CrossRef]
- CDC. SARS (10 Years after). Available online: https://www.cdc.gov/dotw/sars/index.html (accessed on 11 March 2021).
- CDC. MERS in the U.S. Available online: https://www.cdc.gov/coronavirus/mers/us.html (accessed on 12 December 2021).
- Langer, T.; Favarato, M.; Giudici, R.; Bassi, G.; Garberi, R.; Villa, F.; Gay, H.; Zeduri, A.; Bragagnolo, S.; Molteni, A.; et al. Development of machine learning models to predict RT-PCR results for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in patients with influenza-like symptoms using only basic clinical data. Scand. J. Trauma Resusc. Emerg. Med. 2020, 28, 113. [Google Scholar] [CrossRef]











| Samples | Vendor Product | Number of Samples | Label |
|---|---|---|---|
| SARS-CoV-2 | ATCC VR-1986HK [24] | 400 | Positive |
| Blank | NA | 400 | Negative |
| SARS-CoV | ZeptoMetrix NATSARS-ST [25] | 400 | Negative |
| H-CoV OC43 | ZeptoMetrix 0810024CFHI [26] | 400 | Negative |
| MERS-CoV | ZeptoMetrix NATMERS-ST [27] | 400 | Negative |
| H1N1 Influenza A | ZeptoMetrix 0810109CFNHI [28] | 400 | Negative |
| Sample | Number of Samples | Label |
|---|---|---|
| SARS-CoV-2 | 800 | Positive |
| SARS-CoV | 160 | Negative |
| Influenza | 160 | Negative |
| H-CoV | 160 | Negative |
| MERS-COV | 160 | Negative |
| Blank | 160 | Negative |
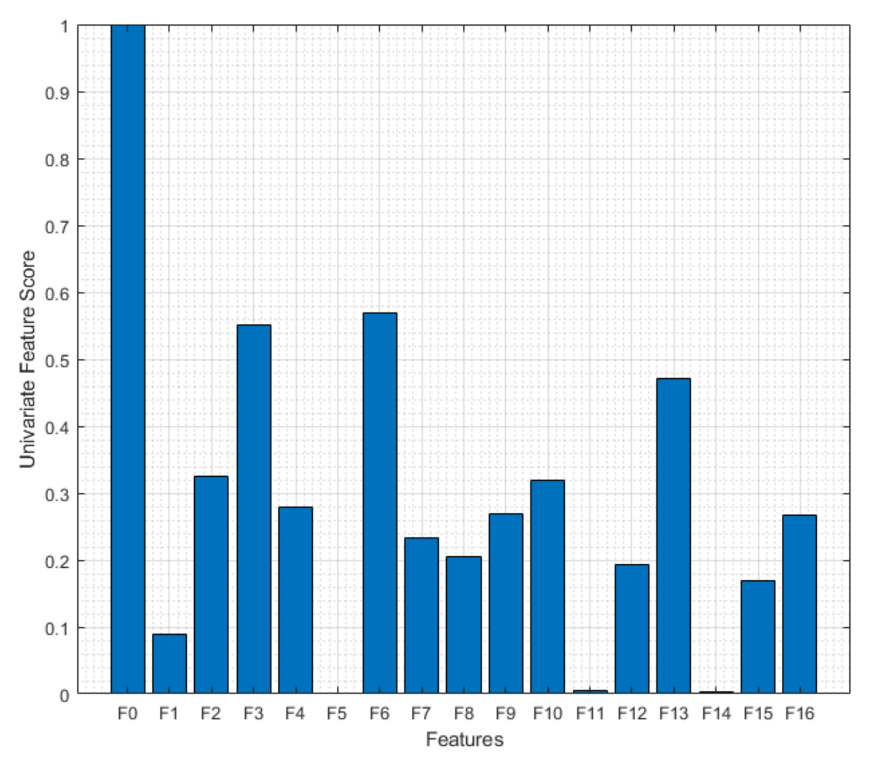
| # | Name | Definition | # | Name | Definition |
|---|---|---|---|---|---|
| F0 | 2% current difference | F9 | Mean absolute deviation | ||
| F1 | Maximum value | F10 | Median absolute deviation | ||
| F2 | Minimum value | F11 | Crest Factor | ||
| F3 | Mean | F12 | Peak2RMS | ||
| F4 | Peak to peak | F13 | Skewness | ||
| F5 | Harmonic mean | F14 | Kurtosis | ||
| F6 | Trimmed mean | Mean excluding outliers | F15 | Shape Factor | |
| F7 | Variance | F16 | RMS | ||
| F8 | Standard deviation | Where | |||
| Eliminated Features | Feature Numbers |
|---|---|
| None Eliminated | F0 F1 F2 F3 F4 F5 F6 F7 F8 F9 F10 F11 F12 F13 F14 F15 F16 |
| Features with 5 Lowest Scores | F0 F2 F3 F4 F6 F7 F8 F9 F10 F13 F16 |
| Features with 10 Lowest Scores | F0 F2 F3 F6 F10 F13 |
| Features with 12 Lowest Scores | F0 F3 F6 F13 |
| Features with 16 Lowest Scores | F0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gecgel, O.; Ramanujam, A.; Botte, G.G. Selective Electrochemical Detection of SARS-CoV-2 Using Deep Learning. Viruses 2022, 14, 1930. https://doi.org/10.3390/v14091930
Gecgel O, Ramanujam A, Botte GG. Selective Electrochemical Detection of SARS-CoV-2 Using Deep Learning. Viruses. 2022; 14(9):1930. https://doi.org/10.3390/v14091930
Chicago/Turabian StyleGecgel, Ozhan, Ashwin Ramanujam, and Gerardine G. Botte. 2022. "Selective Electrochemical Detection of SARS-CoV-2 Using Deep Learning" Viruses 14, no. 9: 1930. https://doi.org/10.3390/v14091930
APA StyleGecgel, O., Ramanujam, A., & Botte, G. G. (2022). Selective Electrochemical Detection of SARS-CoV-2 Using Deep Learning. Viruses, 14(9), 1930. https://doi.org/10.3390/v14091930







