Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19
Abstract
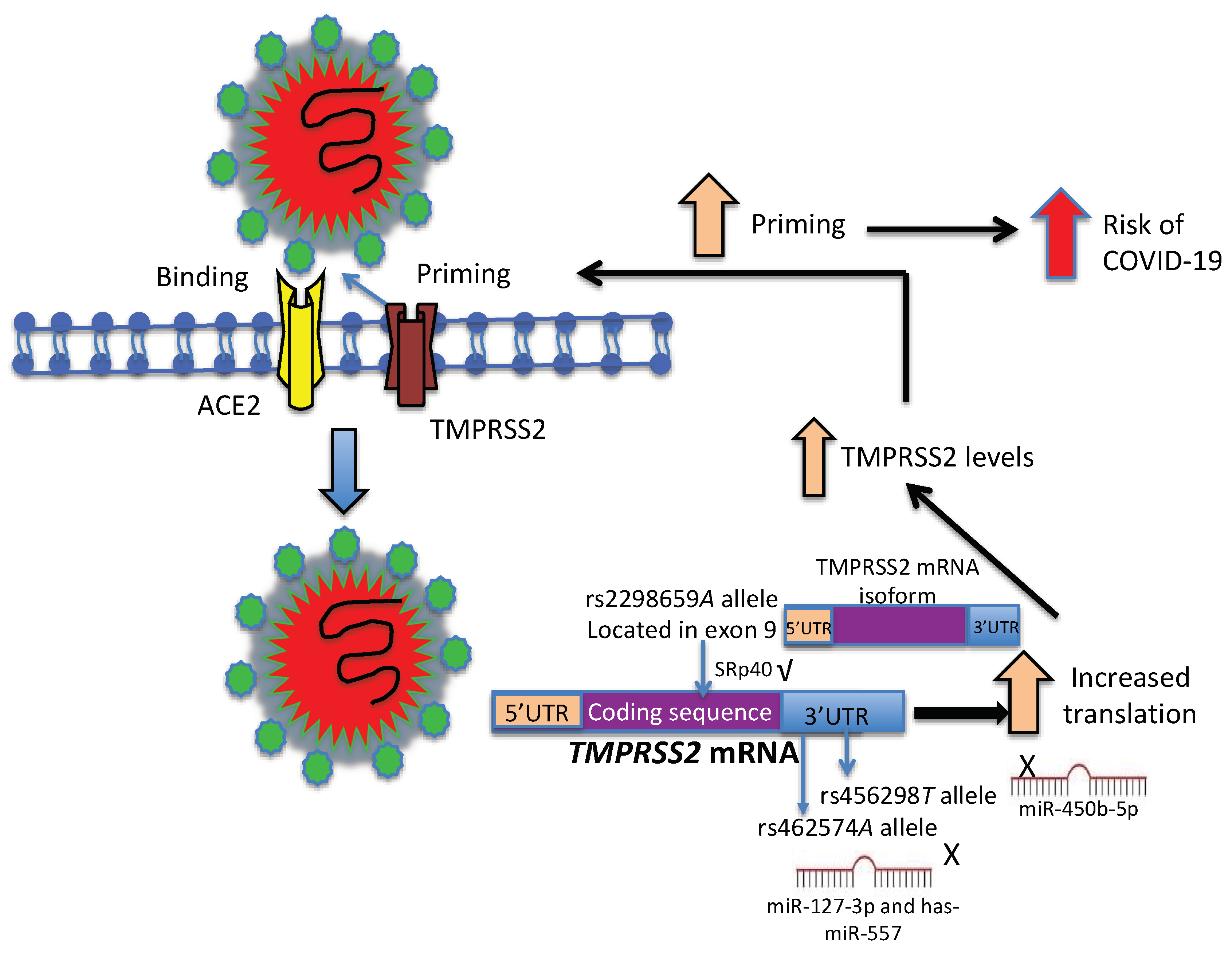
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Sample Handling
2.3. Biochemical Markers
2.4. Genetic Analysis
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the COVID-19 Patients
3.2. Distribution of TMPRSS2 Polymorphisms in COVID-19 Patients and Healthy Controls
3.3. Association of TMPRSS2 Polymorphisms with COVID-19
3.4. Haplotype Analysis
3.5. Biochemical Markers in COVID-19 Patients according to Different Genotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://Covid19.Who.Int (accessed on 28 August 2022).
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.; Herrler, G.; Wu, N.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.; Whittaker, G. Host Cell Entry of Middle East Respiratory Syndrome Coronavirus after Two-Step, Furin-Mediated Activation of the Spike Protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef] [PubMed]
- Gierer, S.; Bertram, S.; Kaup, F.; Wrensch, F.; Heurich, A.; Krämer-Kühl, A.; Welsch, K.; Winkler, M.; Meyer, B.; Drosten, C.; et al. The Spike Protein of the Emerging Betacoronavirus EMC Uses a Novel Coronavirus Receptor for Entry, Can Be Activated by TMPRSS2, and Is Targeted by Neutralizing Antibodies. J. Virol. 2013, 87, 5502–5511. [Google Scholar] [CrossRef]
- Sakai, K.; Ami, Y.; Tahara, M.; Kubota, T.; Anraku, M.; Abe, M.; Nakajima, N.; Sekizuka, T.; Shirato, K.; Suzaki, Y.; et al. The Host Protease TMPRSS2 Plays a Major Role in in Vivo Replication of Emerging H7N9 and Seasonal Influenza Viruses. J. Virol. 2014, 88, 5608–5616. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence That TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [PubMed]
- Tarnow, C.; Engels, G.; Arendt, A.; Schwalm, F.; Sediri, H.; Preuss, A.; Nelson, P.S.; Garten, W.; Klenk, H.-D.; Gabriel, G.; et al. TMPRSS2 Is a Host Factor That Is Essential for Pneumotropism and Pathogenicity of H7N9 Influenza A Virus in Mice. J. Virol. 2014, 88, 4744–4751. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced Isolation of SARS-CoV-2 by TMPRSS2-Expressing Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Biswas, N.; Majumder, P. Analysis of RNA Sequences of 3636 SARS-CoV-2 Collected from 55 Countries Reveals Selective Sweep of One Virus Type. Indian J. Med. Res. 2020, 151, 450–458. [Google Scholar] [CrossRef]
- Gudbjartsson, D.F.; Helgason, A.; Jonsson, H.; Magnusson, O.T.; Melsted, P.; Norddahl, G.L.; Saemundsdottir, J.; Sigurdsson, A.; Sulem, P.; Agustsdottir, A.B.; et al. Spread of SARS-CoV-2 in the Icelandic Population. N. Engl. J. Med. 2020, 382, 2302–2315. [Google Scholar] [CrossRef]
- Suryamohan, K.; Diwanji, D.; Stawiski, E.W.; Gupta, R.; Miersch, S.; Liu, J.; Chen, C.; Jiang, Y.P.; Fellouse, F.A.; Sathirapongsasuti, J.F.; et al. Human ACE2 Receptor Polymorphisms and Altered Susceptibility to SARS-CoV-2. Commun. Biol. 2021, 4, 475. [Google Scholar] [CrossRef]
- Benetti, E.; Tita, R.; Spiga, O.; Ciolfi, A.; Birolo, G.; Bruselles, A.; Doddato, G.; Giliberti, A.; Marconi, C.; Musacchia, F.; et al. ACE2 Gene Variants May Underlie Interindividual Variability and Susceptibility to COVID-19 in the Italian Population. Eur. J. Hum. Genet. 2020, 28, 1602–1614. [Google Scholar] [CrossRef]
- Paniri, A.; Hosseini, M.M.; Akhavan-Niaki, H. First Comprehensive Computational Analysis of Functional Consequences of TMPRSS2 SNPs in Susceptibility to SARS-CoV-2 among Different Populations. J. Biomol. Struct. Dyn. 2021, 39, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Andolfo, I.; Russo, R.; Lasorsa, V.A.; Cantalupo, S.; Rosato, B.E.; Bonfiglio, F.; Frisso, G.; Abete, P.; Cassese, G.M.; Servillo, G.; et al. Common Variants at 21q22.3 Locus Influence MX1 and TMPRSS2 Gene Expression and Susceptibility to Severe COVID-19. iScience 2021, 24, 102322. [Google Scholar] [CrossRef]
- Vargas-Alarcón, G.; Posadas-Sánchez, R.; Ramírez-Bello, J. Variability in Genes Related to SARS-CoV-2 Entry into Host Cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and Its Potential Use in Association Studies. Life Sci. 2020, 260, 118313. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F. New Insights into Genetic Susceptibility of COVID-19: An ACE2 and TMPRSS2 Polymorphism Analysis. BMC Med. 2020, 18, 216. [Google Scholar] [CrossRef] [PubMed]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 Variants and Expression as Candidates to Sex and Country Differences in COVID-19 Severity in Italy. Aging 2020, 12, 10087–10098. [Google Scholar] [CrossRef] [PubMed]
- Smatti, M.M.K.; Al-Sarraj, Y.A.; Albagha, O.; Yassine, H.M.H. Host Genetic Variants Potentially Associated With SARS-CoV-2: A Multi-Population Analysis. Front. Genet. 2020, 11, 578523. [Google Scholar] [CrossRef]
- Elhabyan, A.A.; Elyaacoub, S.; Sanad, E.; Abukhadra, A.; Elhabyan, A.A.; Dinu, V. The Role of Host Genetics in Susceptibility to Severe Viral Infections in Humans and Insights into Host Genetics of Severe COVID-19: A Systematic Review. Virus Res. 2020, 289, 198163. [Google Scholar] [CrossRef]
- Ovsyannikova, I.G.I.; Haralambieva, I.H.I.; Crooke, S.N.S.; Poland, G.A.G.; Kennedy, R.B.R. The Role of Host Genetics in the Immune Response to SARS-CoV-2 and COVID-19 Susceptibility and Severity. Immunol. Rev. 2020, 296, 205–219. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Liu, Y.; Zhang, Z.; Zhai, Y.; Dai, Y.; Wu, Z.; Nie, X.; Du, L. Polymorphisms and Mutations of ACE2 and TMPRSS2 Genes Are Associated with COVID-19: A Systematic Review. Eur. J. Med. Res. 2022, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Irham, L.M.; Chou, W.H.; Calkins, M.J.; Adikusuma, W.; Hsieh, S.L.; Chang, W.C. Genetic Variants That Influence SARS-CoV-2 Receptor TMPRSS2 Expression among Population Cohorts from Multiple Continents. Biochem. Biophys. Res. Commun. 2020, 529, 263–269. [Google Scholar] [CrossRef]
- Latini, A.; Agolini, E.; Novelli, A.; Borgiani, P.; Giannini, R.; Gravina, P.; Smarrazzo, A.; Dauri, M.; Andreoni, M.; Rogliani, P.; et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes 2020, 11, 1010. [Google Scholar] [CrossRef] [PubMed]
- Zarubin, A.; Stepanov, V.; Markov, A.; Kolesnikov, N.; Marusin, A.; Khitrinskaya, I.; Swarovskaya, M.; Litvinov, S.; Ekomasova, N.; Dzhaubermezov, M.; et al. Structural Variability, Expression Profile, and Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy. Genes 2020, 12, 19. [Google Scholar] [CrossRef]
- Wulandari, L.; Hamidah, B.; Pakpahan, C.; Damayanti, N.S.; Kurniati, N.D.; Adiatmaja, C.O.; Wigianita, M.R.; Soedarsono; Husada, D.; Tinduh, D.; et al. Initial Study on TMPRSS2 p.Val160Met Genetic Variant in COVID-19 Patients. Hum. Genom. 2021, 15, 29. [Google Scholar] [CrossRef]
- Schönfelder, K.; Breuckmann, K.; Elsner, C.; Dittmer, U.; Fistera, D.; Herbstreit, F.; Risse, J.; Schmidt, K.; Sutharsan, S.; Taube, C.; et al. Transmembrane Serine Protease 2 Polymorphisms and Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Type 2 Infection: A German Case-Control Study. Front. Genet. 2021, 12, 667231. [Google Scholar] [CrossRef]
- Diamond, R.; Du, K.; Lee, V.; Mohn, K.; Haber, B.; Tewari, D.; Taub, R. Novel Delayed-Early and Highly Insulin-Induced Growth Response Genes. Identification of HRS, a Potential Regulator of Alternative Pre-MRNA Splicing-PubMed. J. Biol. Chem. 1993, 268, 15185–15192. [Google Scholar] [CrossRef]
- Du, K.; Taub, R. Alternative Splicing and Structure of the Human and Mouse SFRS5/HRS/SRp40 Genes. Gene 1997, 204, 243–249. [Google Scholar] [CrossRef]
- Kim, H.R.; Lee, G.O.; Choi, K.H.; Kim, D.K.; Ryu, J.S.; Hwang, K.E.; Na, K.J.; Choi, C.; Kuh, J.H.; Chung, M.J.; et al. SRSF5: A Novel Marker for Small-Cell Lung Cancer and Pleural Metastatic Cancer. Lung Cancer 2016, 99, 57–65. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, R.; Xu, L.; Ma, R.; Xu, L.; Zhu, L.; Hu, J.; An, X. LncRNA Hoxaas3 Promotes Lung Fibroblast Activation and Fibrosis by Targeting MiR-450b-5p to Regulate Runx1. Cell Death Dis. 2020, 11, 706. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; Aggarwal, G.; Wong, J.; Benoit, S.; Vikse, J.; Plebani, M.; Lippi, G. Lactate Dehydrogenase Levels Predict Coronavirus Disease 2019 (COVID-19) Severity and Mortality: A Pooled Analysis. Am. J. Emerg. Med. 2020, 38, 1722–1726. [Google Scholar] [CrossRef]
- Kermali, M.; Khalsa, R.K.; Pillai, K.; Ismail, Z.; Harky, A. The Role of Biomarkers in Diagnosis of COVID-19—A Systematic Review. Life Sci. 2020, 254, 117788. [Google Scholar] [CrossRef]
- Mol, M.B.A.; Strous, M.T.A.; van Osch, F.H.M.; Jeroen Vogelaar, F.; Barten, D.G.; Farchi, M.; Foudraine, N.A.; Gidron, Y. Heart-Rate-Variability (HRV), Predicts Outcomes in COVID-19. PLoS ONE 2021, 16, e0258841. [Google Scholar] [CrossRef] [PubMed]
- McKee, D.L.; Sternberg, A.; Stange, U.; Laufer, S.; Naujokat, C. Candidate Drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020, 157, 104859. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-NCoV) in Vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | COVID-19 Patients (n = 609) |
|---|---|
| Age (years) | 50.3 ± 14.56 |
| Sex male n, (%) | 353, (58) |
| Temperature (°C) | 36.7 ± 0.97 |
| Oxygen saturation (SpO2) | 86.85 ± 11.28 |
| Heart rate (bpm) | 90.94 ± 20.15 |
| Comorbidities | |
| Obesity n (%) | 293 (47.7) |
| TDM2 n (%) | 162 (26.3) |
| Hypertension n (%) | 174 (28.3) |
| Biochemical markers | |
| Creatinine (mg/dL) | 0.86 [0.68–1.16] |
| Ferritin (ng/µL) | 448 [216–931.7] |
| LDH (U/dL) | 305 [219–406] |
| Protein C Reactive (mg/dL) | 14 [2.89–54.2] |
| Total bilirubin (mg/dL) | 1.0 [0.44–0.87] |
| ALT (U/dL) | 36 [21.1–61.1] |
| AST (U/dL) | 38 [24–81] |
| Hemoglobin (g/dL) | 14 [12.1–15.6] |
| Platelets (109/L) | 257 [199–326] |
| Genotype Frequency | MAF | Model | OR (95%CI) | p | |||
|---|---|---|---|---|---|---|---|
| rs12329760 | CC | CT | TT | T | |||
| Controls (n = 291) n | 0.735 214 | 0.241 70 | 0.024 7 | 0.144 | Co-dominant1 Co-dominant2 | 1.15 (0.77–1.71) 0.86 (0.28–2.69) | 0.982 0.749 |
| Dominant | 1.12 (0.77–1.64) | 0.549 | |||||
| COVID-19 (n = 609) | 0.741 | 0.239 | 0.020 | 0.140 | Recessive | 0.83 (0.27–2.58) | 0.751 |
| n | 451 | 146 | 12 | Heterozygote | 1.16 (0.78–1.71) | 0.471 | |
| Additive | 1.08 (0.77–1.51) | 0.670 | |||||
| rs2298659 | GG | GA | AA | A | |||
| Controls (n = 291) n | 0.577 168 | 0.375 109 | 0.048 14 | 0.235 | Co-dominant1 Co-dominant2 | 1.13 (0.79–1.63) 2.61 (1.30–5.23) | 0.475 0.018 |
| Dominant | 1.31 (0.93–1.84) | 0.129 | |||||
| COVID-19 (n = 607) | 0.514 | 0.376 | 0.110 | 0.298 | Recessive | 2.49 (1.26–4.90) | 0.006 |
| n | 312 | 228 | 67 | Heterozygote | 1.00 (0.70–1.43) | 0.978 | |
| Additive | 1.38 (1.05–1.80) | 0.019 | |||||
| rs456298 | AA | AT | TT | T | |||
| Controls (n = 291) n | 0.354 103 | 0.464 135 | 0.182 53 | 0.414 | Co-dominant1 Co-dominant2 | 1.51 (1.09–2.09) 2.27 (1.40–3.69) | 0.014 0.004 |
| Dominant | 1.65 (1.13–2.42) | 0.009 | |||||
| COVID-19 (n = 609) | 0.245 | 0.486 | 0.269 | 0.512 | Recessive | 1.82 (1.20–2.74) | 0.004 |
| n | 149 | 296 | 164 | Heterozygote | 0.99 (0.70–1.39) | 0.939 | |
| Additive | 1.50 (1.18–1.91) | 0.0009 | |||||
| rs462574 | GG | GA | AA | A | |||
| Controls (n = 291) n | 0.485 141 | 0.419 122 | 0.096 28 | 0.306 | Co-dominant1 Co-dominant | 1.46 (1.07–1.96) 2.46 (1.42–4.27) | 0.017 0.004 |
| Dominant | 1.43 (1.02–2.02) | 0.041 | |||||
| COVID-19 (n = 609) | 0.361 | 0.455 | 0.184 | 0.411 | Recessive | 2.24 (1.33–3.17) | 0.002 |
| n | 220 | 277 | 112 | Heterozygote | 0.97 (0.69–1.37) | 0.879 | |
| Additive | 1.46 (1.14–1.87) | 0.003 | |||||
| Haplotype | COVID-19 (n = 607) | Controls (n = 291) | OR | 95%CI | p |
|---|---|---|---|---|---|
| Hf (n) | Hf (n) | ||||
| ATGC | 0.372 (226) | 0.289 (84) | 1.51 | 1.12–2.05 | 0.006 |
| GAGC | 0.278 (169) | 0.430 (125) | 0.52 | 0.39–0.70 | <0.001 |
| GAAC | 0.160 (97) | 0.110 (32) | 1.58 | 1.03–2.42 | 0.017 |
| GTAT | 0.091 (55) | 0.096 (28) | 0.95 | 0.59–1.54 | 0.860 |
| GAAT | 0.023 (14) | 0.017 (5) | 1.38 | 0.49–3.87 | 0.538 |
| GTGC | 0.016 (10) | 0.014 (4) | 1.22 | 0.38–3.95 | 0.729 |
| GAGT | 0.008 (5) | 0.024 (7) | 0.34 | 0.10–1.09 | 0.029 |
| AAGC | 0.013 (8) | 0.003 (1) | 3.95 | 0.49–31.8 | 0.081 |
| rs462574 | Genotypes | p | ||
|---|---|---|---|---|
| GG | GA | AA | ||
| Creatinine (mg/dL) | 1.35 ± 1.76 | 1.36 ± 2.51 | 1.54 ± 1.89 | 0.801 |
| Ferritin (ng/μL) | 612 ± 619.4 | 624 ± 588.4 | 764 ± 801 | 0.208 |
| LDH (U/dL) | 308 ± 142.6 | 352 ± 181.6 | 380.2 ± 195.7 | 0.005 |
| C reactive protein (mg/dL) | 49.4 ± 85.4 | 51.4 ± 83.9 | 63.2 ± 103.5 | 0.537 |
| Total bilirubin (mg/dL) | 1.06 ± 2.1 | 0.87 ± 1.21 | 0.80 ± 0.84 | 0.413 |
| ALT (U/dL) | 44.6 ± 39.2 | 53.9 ± 77.2 | 52.7 ± 47.9 | 0.393 |
| AST (U/dL) | 45.6 ± 32.6 | 51 ± 74.1 | 53.8 ± 41.3 | 0.562 |
| Hemoglobin (g/dL) | 13.8 ± 2.96 | 13.7 ± 2.63 | 13.6 ± 2.74 | 0.876 |
| Platelets (10−6/µL) | 269 ± 115 | 297 ± 136 | 280 ± 124 | 0.113 |
| Heart rate (bpm) | 88 ± 19.4 | 92 ± 20.5 | 94.6 ± 20 | 0.028 |
| Temperature (°C) | 36.5 ± 0.86 | 36.7 ± 0.74 | 37 ± 1.52 | 0.037 |
| Oxygen saturation (SpO2) | 87 ± 11.9 | 87 ± 11.42 | 87 ± 9.81 | 0.999 |
| rs456298 | ||||
| AA | AT | TT | ||
| Creatinine (mg/dL) | 1.46 ± 2.02 | 1.26 ± 2.30 | 1.54 ± 2.04 | 0.517 |
| Ferritin (ng/µL) | 632 ± 640.9 | 623 ± 588.5 | 696 ± 737 | 0.560 |
| LDH (U/dL) | 314.9 ± 142.9 | 336 ± 164.4 | 377 ± 206.3 | 0.020 |
| C reactive protein (mg/dL) | 74.1 ± 82.8 | 51.6 ± 85.5 | 59.5 ± 96.2 | 0.132 |
| Total bilirubin (mg/dL) | 1.28 ± 2.56 | 0.82 ± 1.06 | 0.79 ± 0.76 | 0.032 |
| ALT (U/dL) | 44.1 ± 40.8 | 54.3 ± 76.9 | 49.7 ± 43.8 | 0.416 |
| AST (U/dL) | 46.9 ± 35.5 | 51 ± 73.2 | 50.1 ± 38.8 | 0.850 |
| Hemoglobin (g/dL) | 14 ± 3.00 | 14 ± 2.65 | 13.5 ± 2.74 | 0.274 |
| Platelets (10−6/μL) | 269.0 ± 123.5 | 291 ± 132.7 | 284 ± 119.4 | 0.345 |
| Heart rate (bpm) | 89.6 ± 19.4 | 90 ± 20.0 | 94.4 ± 20.1 | 0.086 |
| Temperature (ºC) | 36.6 ± 0.80 | 36.6 ± 0.79 | 37 ± 1.33 | 0.001 |
| Oxygen saturation (SpO2) | 87 ± 12.04 | 87 ± 11.5 | 87 ± 10.3 | 0.999 |
| rs2298659 | ||||
| GG | GA | AA | ||
| Creatinine (mg/dL) | 1.45 ± 2.14 | 1.26 ± 2.24 | 1.54 ± 2.14 | 0.642 |
| Ferritin (ng/µL) | 702 ± 742.6 | 601 ± 539.2 | 536 ± 478.9 | 0.174 |
| LDH (U/dL) | 342 ± 171.2 | 345 ± 185.1 | 334 ± 152.7 | 0.930 |
| C reactive protein (mg/dL) | 50 ± 82.9 | 58.9 ± 97.2 | 50.1 ± 79.3 | 0.634 |
| Total bilirubin (mg/dL) | 0.90 ± 1.29 | 0.90 ± 1.23 | 1.04 ± 2.79 | 0.846 |
| ALT (U/dL) | 48.4 ± 43.42 | 54.9 ± 85.5 | 45.2 ± 39.4 | 0.527 |
| AST (U/dL) | 49.4 ± 39.3 | 52.3 ± 81.9 | 42.6 ± 26.4 | 0.619 |
| Hemoglobin (g/dL) | 13.7 ± 2.83 | 13.68 ± 2.59 | 14.1 ± 2.99 | 0.658 |
| Platelets (10−6/µL) | 277 ± 125.9 | 292 ± 121.7 | 292 ± 147.7 | 0.467 |
| Heart rate (bpm) | 92 ± 20.4 | 89 ± 19.1 | 91 ± 21.5 | 0.314 |
| Temperature (°C) | 36.7 ± 1.09 | 36.6 ± 0.79 | 37 ± 0.92 | 0.038 |
| Oxygen saturation (SpO2) | 87 ± 10.82 | 88 ± 11.8 | 84 ± 11.8 | 0.086 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Posadas-Sánchez, R.; Fragoso, J.M.; Sánchez-Muñoz, F.; Rojas-Velasco, G.; Ramírez-Bello, J.; López-Reyes, A.; Martínez-Gómez, L.E.; Sierra-Fernández, C.; Rodríguez-Reyna, T.; Regino-Zamarripa, N.E.; et al. Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19. Viruses 2022, 14, 1976. https://doi.org/10.3390/v14091976
Posadas-Sánchez R, Fragoso JM, Sánchez-Muñoz F, Rojas-Velasco G, Ramírez-Bello J, López-Reyes A, Martínez-Gómez LE, Sierra-Fernández C, Rodríguez-Reyna T, Regino-Zamarripa NE, et al. Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19. Viruses. 2022; 14(9):1976. https://doi.org/10.3390/v14091976
Chicago/Turabian StylePosadas-Sánchez, Rosalinda, José Manuel Fragoso, Fausto Sánchez-Muñoz, Gustavo Rojas-Velasco, Julian Ramírez-Bello, Alberto López-Reyes, Laura E. Martínez-Gómez, Carlos Sierra-Fernández, Tatiana Rodríguez-Reyna, Nora Elemi Regino-Zamarripa, and et al. 2022. "Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19" Viruses 14, no. 9: 1976. https://doi.org/10.3390/v14091976
APA StylePosadas-Sánchez, R., Fragoso, J. M., Sánchez-Muñoz, F., Rojas-Velasco, G., Ramírez-Bello, J., López-Reyes, A., Martínez-Gómez, L. E., Sierra-Fernández, C., Rodríguez-Reyna, T., Regino-Zamarripa, N. E., Ramírez-Martínez, G., Zuñiga-Ramos, J., & Vargas-Alarcón, G. (2022). Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19. Viruses, 14(9), 1976. https://doi.org/10.3390/v14091976









