A Simple and Effective Method to Concentrate Hepatitis C Virus: Aqueous Two-Phase System Allows Highly Efficient Enrichment of Enveloped Viruses
Abstract
:1. Introduction
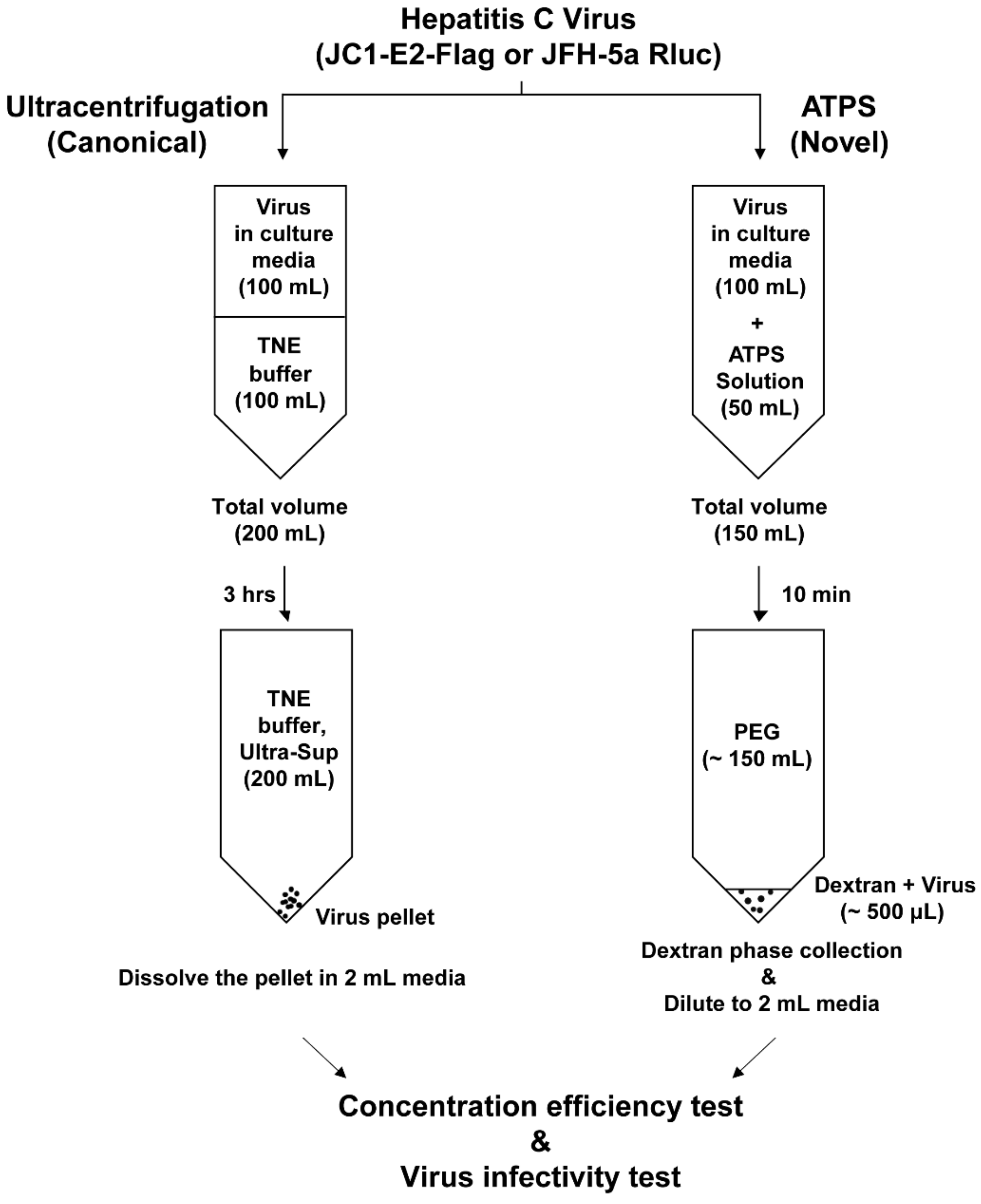
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Virus Production
2.3. Ultracentrifugation
2.4. Aqueous Two-Phase System (ATPS)
2.5. Characterization of Viruses
2.6. Quantification of HCV RNA
2.7. Immunocytochemistry
2.8. Measurement of Virus Infectivity
2.9. Efficiency of HCV Recovery
3. Results
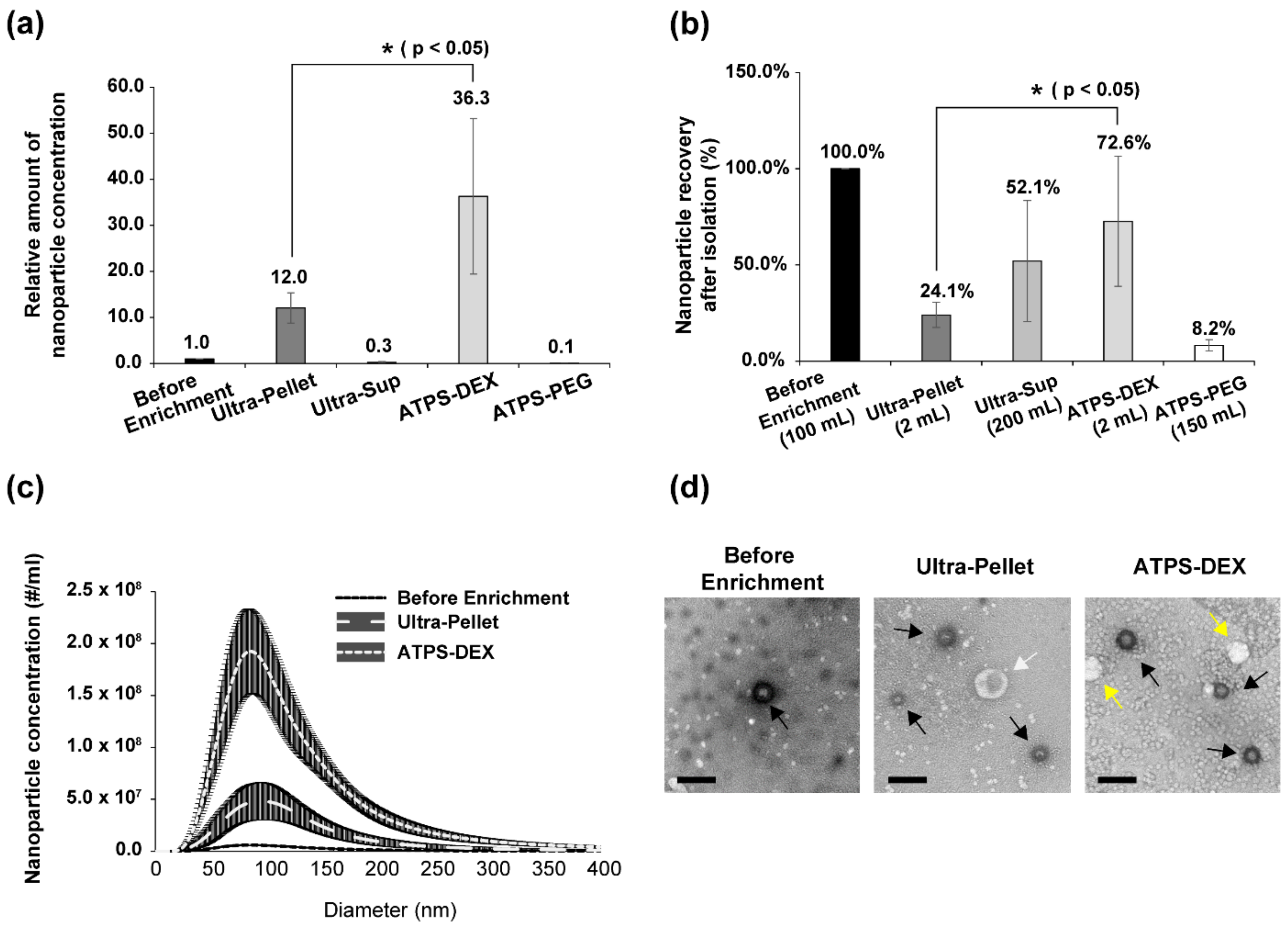
3.1. ATPS Is a Highly Effective Virus Concentration System (Comparison between ATPS and Ultracentrifugation)
3.2. The Size Distribution of Nanoparticles Enriched by ATPS and Ultracentrifugation
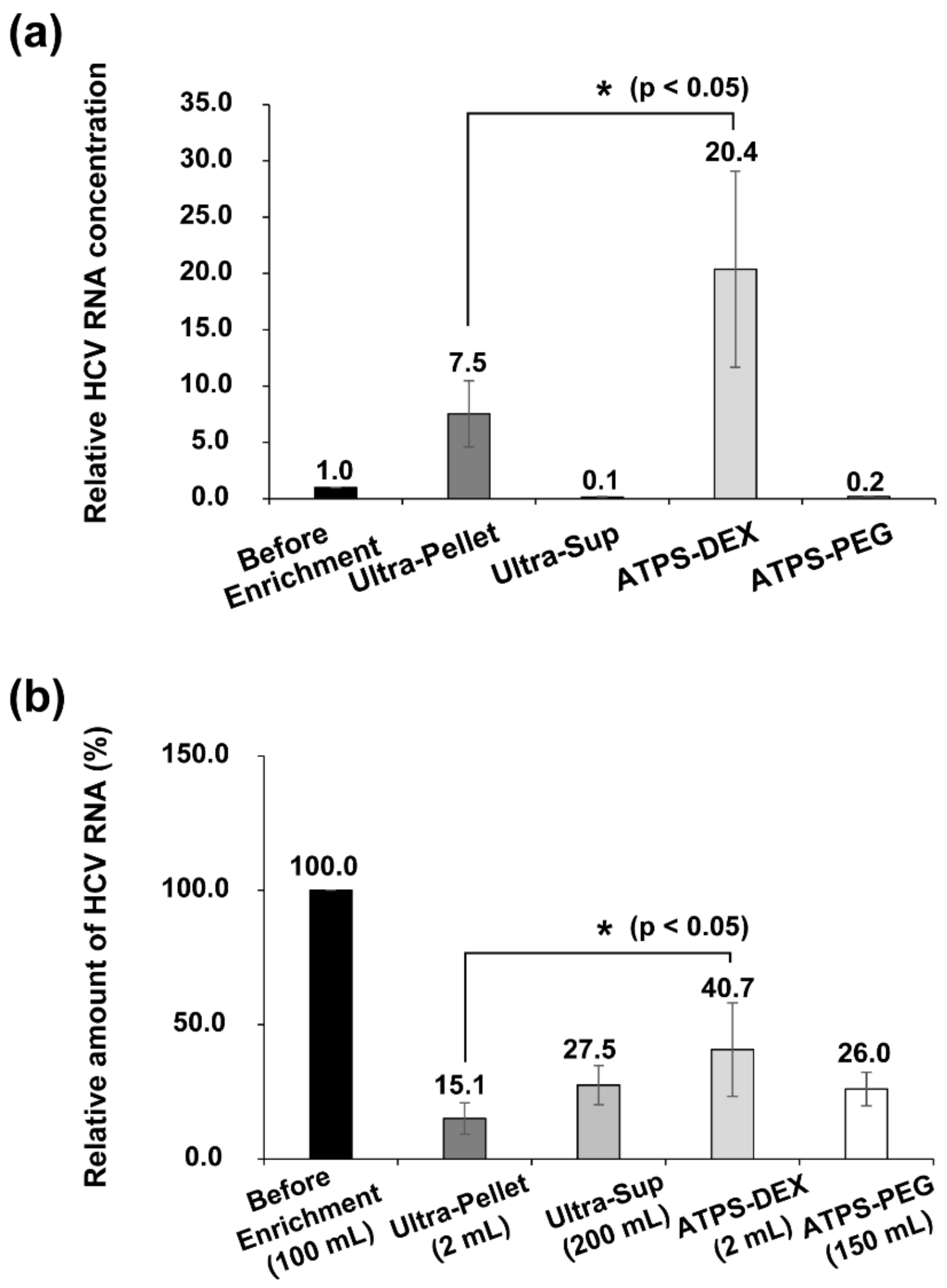
3.3. Quantification of HCV RNAs before and after Enrichment Processes
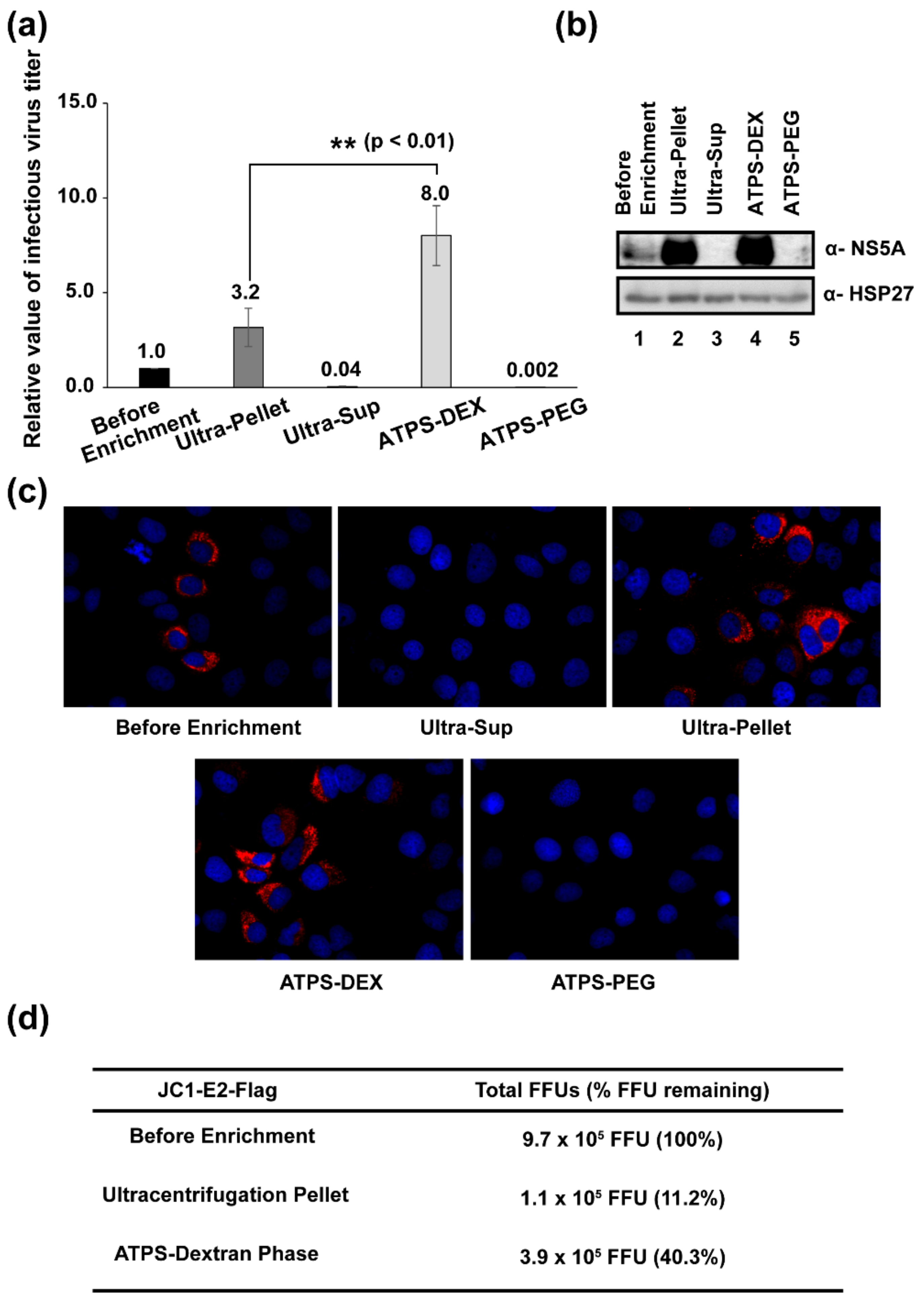
3.4. Determination of Viral Titers before and after Enrichment Processes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morenweiser, R. Downstream processing of viral vectors and vaccines. Gene Ther. 2005, 12, S103–S110. [Google Scholar] [CrossRef] [PubMed]
- Keum, S.J.; Park, S.M.; Park, J.H.; Jung, J.H.; Shin, E.J.; Jang, S.K. The specific infectivity of hepatitis C virus changes through its life cycle. Virology 2012, 433, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Kutner, R.H.; Zhang, X.-Y.; Reiser, J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009, 4, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Szelechowski, M.; Bergeron, C.; Gonzalez-Dunia, D.; Klonjkowski, B. Production and purification of non replicative canine adenovirus type 2 derived vectors. JoVE J. Vis. Exp. 2013, 82, e50833. [Google Scholar] [CrossRef] [PubMed]
- Nasukawa, T.; Uchiyama, J.; Taharaguchi, S.; Ota, S.; Ujihara, T.; Matsuzaki, S.; Murakami, H.; Mizukami, K.; Sakaguchi, M. Virus purification by CsCl density gradient using general centrifugation. Arch. Virol. 2017, 162, 3523–3528. [Google Scholar] [CrossRef] [PubMed]
- Reiser, J. Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 2000, 7, 910–913. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020, 739, 139960. [Google Scholar] [CrossRef]
- Bartolomé, J.; López-Alcorocho, J.M.; Castillo, I.; Rodríguez-Iñigo, E.; Quiroga, J.A.; Palacios, R.; Carreno, V. Ultracentrifugation of serum samples allows detection of hepatitis C virus RNA in patients with occult hepatitis C. J. Virol. 2007, 81, 7710–7715. [Google Scholar] [CrossRef]
- Dustin, L.B.; Bartolini, B.; Capobianchi, M.R.; Pistello, M. Hepatitis C virus: Life cycle in cells, infection and host response, and analysis of molecular markers influencing the outcome of infection and response to therapy. Clin. Microbiol. Infect. 2016, 22, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Holmes, J.A.; Rutledge, S.M.; Chung, R.T. Direct-acting antiviral treatment for hepatitis C. Lancet 2019, 393, 1392–1394. [Google Scholar] [CrossRef]
- Manns, M.P.; Buti, M.; Gane, E.; Pawlotsky, J.-M.; Razavi, H.; Terrault, N.; Younossi, Z. Hepatitis C virus infection. Nat. Rev. Dis. Primers 2017, 3, 17006. [Google Scholar] [CrossRef] [PubMed]
- Bassendine, M.F.; Sheridan, D.A.; Felmlee, D.J.; Bridge, S.H.; Toms, G.L.; Neely, R.D.G. HCV and the hepatic lipid pathway as a potential treatment target. J. Hepatol. 2011, 55, 1428–1440. [Google Scholar] [CrossRef] [PubMed]
- Catanese, M.T.; Uryu, K.; Kopp, M.; Edwards, T.J.; Andrus, L.; Rice, W.J.; Silvestry, M.; Kuhn, R.J.; Rice, C.M. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl. Acad. Sci. USA 2013, 110, 9505–9510. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.-A.; Lièvre, M.; Peyrol, S.; De Sequeira, S.; Berthillon, P.; Ruigrok, R.W.; Trépo, C. Enveloped particles in the serum of chronic hepatitis C patients. Virology 2005, 336, 144–153. [Google Scholar] [CrossRef]
- Ammersbach, M.; Bienzle, D. Methods for assessing feline immunodeficiency virus infection, infectivity and purification. Vet. Immunol. Immunopathol. 2011, 143, 202–214. [Google Scholar] [CrossRef]
- Jiang, W.; Hua, R.; Wei, M.; Li, C.; Qiu, Z.; Yang, X.; Zhang, C. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci. Rep. 2015, 5, 13875. [Google Scholar] [CrossRef]
- Prado, T.; Guilayn, W.d.C.P.B.; Gaspar, A.M.C.; Miagostovich, M.P. The efficiency of concentration methods used to detect enteric viruses in anaerobically digested sludge. Mem. Do Inst. Oswaldo Cruz 2013, 108, 77–83. [Google Scholar] [CrossRef]
- Ye, Y.; Ellenberg, R.M.; Graham, K.E.; Wigginton, K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016, 50, 5077–5085. [Google Scholar] [CrossRef]
- Shin, H.; Han, C.; Labuz, J.M.; Kim, J.; Kim, J.; Cho, S.; Gho, Y.S.; Takayama, S.; Park, J. High-yield isolation of extracellular vesicles using aqueous two-phase system. Sci. Rep. 2015, 5, 13103. [Google Scholar] [CrossRef]
- Kim, J.; Shin, H.; Kim, J.; Kim, J.; Park, J. Isolation of High-Purity Extracellular Vesicles by Extracting Proteins Using Aqueous Two-Phase System. PLoS ONE 2015, 10, e0129760. [Google Scholar]
- Kırbaş, O.K.; Bozkurt, B.T.; Asutay, A.B.; Mat, B.; Ozdemir, B.; Öztürkoğlu, D.; Ölmez, H.; İşlek, Z.; Şahin, F.; Taşlı, P.N. Optimized Isolation of Extracellular Vesicles From Various Organic Sources Using Aqueous Two-Phase System. Sci. Rep. 2019, 9, 19159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merz, A.; Long, G.; Hiet, M.-S.; Brügger, B.; Chlanda, P.; Andre, P.; Wieland, F.; Krijnse-Locker, J.; Bartenschlager, R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 2011, 286, 3018–3032. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.S.; Keum, S.J.; Jang, S.K. Generation of a cell culture-adapted hepatitis C virus with longer half life at physiological temperature. PLoS ONE 2011, 6, e22808. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Kaito, M.; Ishida, S.; Tanaka, H.; Horiike, S.; Fujita, N.; Adachi, Y.; Kohara, M.; Konishi, M.; Watanabe, S. Morphology of hepatitis C and hepatitis B virus particles as detected by immunogold electron microscopy. Med. Mol. Morphol. 2006, 39, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.; Coumans, F. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1555419. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, N.; Bowman, W.P.; Lacko, A.G. Lipoprotein based drug delivery: Potential for pediatric cancer applications. World J. Pharmacol. 2015, 4, 172–179. [Google Scholar] [CrossRef]
- Lauer, M.E.; Graff-Meyer, A.; Rufer, A.C.; Maugeais, C.; von der Mark, E.; Matile, H.; D’Arcy, B.; Magg, C.; Ringler, P.; Müller, S.A. Cholesteryl ester transfer between lipoproteins does not require a ternary tunnel complex with CETP. J. Struct. Biol. 2016, 194, 191–198. [Google Scholar] [CrossRef]
- Negrete, A.; Ling, T.C.; Lyddiatt, A. Production of adenoviral vectors and its recovery. Process Biochem. 2007, 42, 1107–1113. [Google Scholar] [CrossRef]
- Gomes, G.A.; Azevedo, A.M.; Aires-Barros, M.R.; Prazeres, D.M.F. Purification of plasmid DNA with aqueous two phase systems of PEG 600 and sodium citrate/ammonium sulfate. Sep. Purif. Technol. 2009, 65, 22–30. [Google Scholar] [CrossRef]
- Effio, C.L.; Wenger, L.; Ötes, O.; Oelmeier, S.A.; Kneusel, R.; Hubbuch, J. Downstream processing of virus-like particles: Single-stage and multi-stage aqueous two-phase extraction. J. Chromatogr. A 2015, 1383, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Park, Y.H.; Kim, Y.-G.; Lee, J.Y.; Park, J. Aqueous two-phase system to isolate extracellular vesicles from urine for prostate cancer diagnosis. PLoS ONE 2018, 13, e0194818. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Yi, J.; Yu, J.; Park, J.; Jang, S.K. A Simple and Effective Method to Concentrate Hepatitis C Virus: Aqueous Two-Phase System Allows Highly Efficient Enrichment of Enveloped Viruses. Viruses 2022, 14, 1987. https://doi.org/10.3390/v14091987
Kim H, Yi J, Yu J, Park J, Jang SK. A Simple and Effective Method to Concentrate Hepatitis C Virus: Aqueous Two-Phase System Allows Highly Efficient Enrichment of Enveloped Viruses. Viruses. 2022; 14(9):1987. https://doi.org/10.3390/v14091987
Chicago/Turabian StyleKim, Heesun, Johan Yi, Jinbae Yu, Jaesung Park, and Sung Key Jang. 2022. "A Simple and Effective Method to Concentrate Hepatitis C Virus: Aqueous Two-Phase System Allows Highly Efficient Enrichment of Enveloped Viruses" Viruses 14, no. 9: 1987. https://doi.org/10.3390/v14091987
APA StyleKim, H., Yi, J., Yu, J., Park, J., & Jang, S. K. (2022). A Simple and Effective Method to Concentrate Hepatitis C Virus: Aqueous Two-Phase System Allows Highly Efficient Enrichment of Enveloped Viruses. Viruses, 14(9), 1987. https://doi.org/10.3390/v14091987





