Continuous Circulation of Yellow Fever among Rural Populations in the Central African Republic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Detection
2.2. Whole-Genome Sequencing
2.3. Bioinformatics and Phylogenetic Analysis
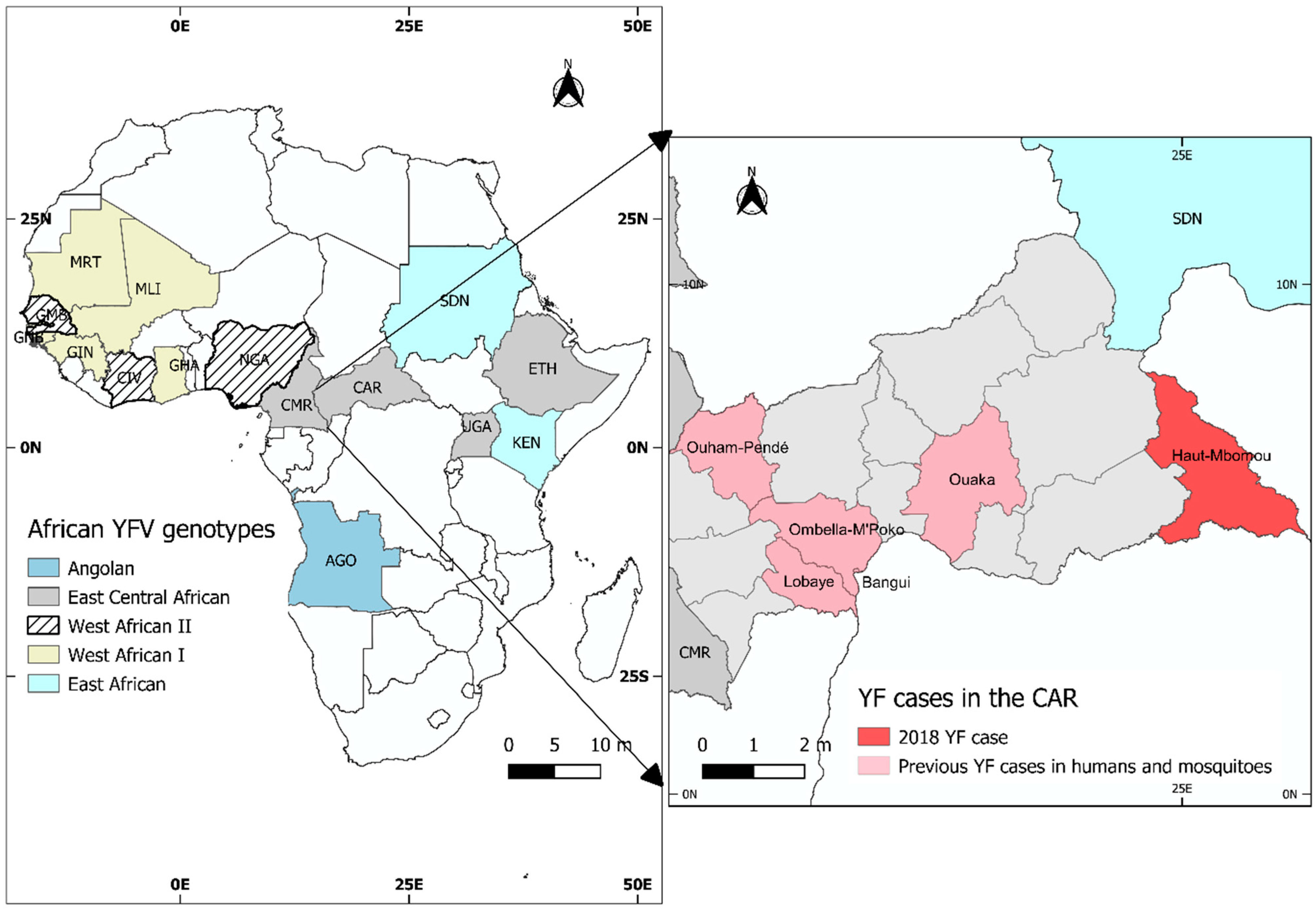
3. Results
3.1. Case Description
3.2. Molecular Analysis of the YFV Strain Identified
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Yellow Fever. 7 May 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/yellow-fever (accessed on 13 May 2021).
- Shearer, F.M.; Moyes, C.L.; Pigott, D.M.; Brady, O.J.; Marinho, F.; Deshpande, A.; Longbottom, J.; Browne, A.J.; Kraemer, M.U.G.; O’Reilly, K.M.; et al. Global yellow fever vaccination coverage from 1970 to 2016: An adjusted retrospective analysis. Lancet Infect. Dis. 2017, 17, 1209–1217. [Google Scholar] [CrossRef]
- Gaythorpe, K.A.; Hamlet, A.; Jean, K.; Garkauskas Ramos, D.; Cibrelus, L.; Garske, T.; Ferguson, N. The global burden of yellow fever. Elife 2021, 10, e64670. [Google Scholar] [CrossRef] [PubMed]
- Beasley, D.W.; McAuley, A.J.; Bente, D.A. Yellow fever virus: Genetic and phenotypic diversity and implications for detection, prevention and therapy. Antivir. Res. 2015, 115, 48–70. [Google Scholar] [CrossRef]
- Sacchetto, L.; Drumond, B.P.; Han, B.A.; Nogueira, M.L.; Vasilakis, N. Re-emergence of yellow fever in the neotropics-quo vadis? Emerg. Top. Life Sci. 2020, 4, 399–410. [Google Scholar]
- Mutebi, J.P.; Wang, H.; Li, L.; Bryant, J.E.; Barrett, A.D. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. J. Virol. 2001, 75, 6999–7008. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Vasconcelos, P.F. Yellow fever. J. Clin. Virol. 2015, 64, 160–173. [Google Scholar] [CrossRef]
- Grobbelaar, A.A.; Weyer, J.; Moolla, N.; Jansen van Vuren, P.; Moises, F.; Paweska, J.T. Resurgence of Yellow Fever in Angola, 2015–2016. Emerg. Infect. Dis. 2016, 22, 1854–1855. [Google Scholar] [CrossRef]
- Tuboi, S.H.; Costa, Z.G.; da Costa Vasconcelos, P.F.; Hatch, D. Clinical and epidemiological characteristics of yellow fever in Brazil: Analysis of reported cases 1998–2002. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 169–175. [Google Scholar] [CrossRef]
- Ho, Y.L.; Joelsons, D.; Leite, G.F.C.; Malbouisson, L.M.S.; Song, A.T.W.; Perondi, B.; Andrade, L.C.; Pinto, L.F.; D’Albuquerque, L.A.C.; Segurado, A.A.C. Severe yellow fever in Brazil: Clinical characteristics and management. J. Travel. Med. 2019, 26, taz040. [Google Scholar] [CrossRef]
- Alhakimi, H.A.; Mohamed, O.G.; Khogaly, H.S.E.; Arafa, K.A.O.; Ahmed, W.A. Epidemiological, Clinical and Entomological Characteristics of Yellow Fever Outbreak in Darfur 2012. AIMS Public Health 2015, 2, 132–141. [Google Scholar] [CrossRef]
- Servadio, J.L.; Muñoz-Zanzi, C.; Convertino, M. Estimating case fatality risk of severe Yellow Fever cases: Systematic literature review and meta-analysis. BMC Infect. Dis. 2021, 21, 819. [Google Scholar] [CrossRef] [PubMed]
- Mathiot, C.C.; Gonzalez, J.P.; Georges, A.J. Current problems of arboviruses in central Africa. Bull. Soc. Pathol. Exot. Fil. 1988, 81, 396–401. [Google Scholar]
- Staples, J.E.; Diallo, M.; Janusz, K.B.; Manengu, C.; Lewis, R.F.; Perea, W.; Yactayo, S.; Sall, A.A. Yellow fever risk assessment in the Central African Republic. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 608–615. [Google Scholar] [CrossRef]
- WHO. Yellow Fever in the Central African Republic. 2009. Available online: https://www.who.int/csr/don/2009_12_01/en/ (accessed on 13 May 2021).
- Rachas, A.; Nakoune, E.; Bouscaillou, J.; Paireau, J.; Selekon, B.; Senekian, D.; Fontanet, A.; Kazanji, M. Timeliness of yellow fever surveillance, Central African Republic. Emerg. Infect. Dis. 2014, 20, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
- Ngoagouni, C.; Kamgang, B.; Manirakiza, A.; Nangouma, A.; Paupy, C.; Nakoune, E.; Kazanji, M. Entomological profile of yellow fever epidemics in the Central African Republic, 2006–2010. Parasit. Vector. 2012, 5, 175. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Patel, P.; Yillah, J.; Weidmann, M.; Méndez, J.A.; Nakouné, E.R.; Niedrig, M. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J. Clin. Microbiol. 2012, 50, 4054–4060. [Google Scholar] [CrossRef]
- Weidmann, M.; Sanchez-Seco, M.P.; Sall, A.A.; Ly, P.O.; Thiongane, Y.; Lô, M.M.; Schley, H.; Hufert, F.T. Rapid detection of important human pathogenic Phleboviruses. J. Clin. Virol. 2008, 41, 138–142. [Google Scholar] [CrossRef]
- Weidmann, M.; Sall, A.A.; Manuguerra, J.C.; Koivogui, L.; Adjami, A.; Traoré, F.F.; Hedlund, K.O.; Lindegren, G.; Mirazimi, A. Quantitative analysis of particles, genomes and infectious particles in supernatants of haemorrhagic fever virus cell cultures. Virol. J. 2011, 8, 8–81. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Method. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, M.R.; Palacios, G.; Cardoso, J.F.; Martins, L.C.; Sousa, E.C., Jr.; de Lima, C.P.; Medeiros, D.B.; Savji, N.; Desai, A.; Rodrigues, S.G.; et al. Genomic and phylogenetic characterization of Brazilian yellow fever virus strains. J. Virol. 2012, 86, 13263–13271. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- McMullan, L.K.; Frace, M.; Sammons, S.A.; Shoemaker, T.; Balinandi, S.; Wamala, J.F.; Lutwama, J.J.; Downing, R.G.; Stroeher, U.; MacNeil, A.; et al. Using next generation sequencing to identify yellow fever virus in Uganda. Virology 2012, 422, 1–5. [Google Scholar] [CrossRef]
- Sall, A.A.; Faye, O.; Diallo, M.; Firth, C.; Kitchen, A.; Holmes, E.C. Yellow fever virus exhibits slower evolutionary dynamics than dengue virus. J. Virol. 2010, 84, 765–772. [Google Scholar] [CrossRef]
- Mulchandani, R.; Massebo, F.; Bocho, F.; Jeffries, C.L.; Walker, T.; Messenger, L.A. A community-level investigation following a yellow fever virus outbreak in South Omo Zone, South-West Ethiopia. PeerJ 2019, 7, e6466. [Google Scholar] [CrossRef]
- Sanders, E.J.; Marfin, A.A.; Tukei, P.M.; Kuria, G.; Ademba, G.; Agata, N.N.; Ouma, J.O.; Cropp, C.B.; Karabatsos, N.; Reiter, P.; et al. First recorded outbreak of yellow fever in Kenya, 1992–1993. I. Epidemiologic investigations. Am. J. Trop. Med. Hyg. 1998, 59, 644–649. [Google Scholar] [CrossRef]
- Phalkey, R.K.; Yamamoto, S.; Awate, P.; Marx, M. Challenges with the implementation of an Integrated Disease Surveillance and Response (IDSR) system: Systematic review of the lessons learned. Health Policy Plan. 2015, 30, 131–143. [Google Scholar] [CrossRef]
- Joseph Wu, T.S.; Kagoli, M.; Kaasboll, J.J.; Bjune, G.A. Integrated Disease Surveillance and Response (IDSR) in Malawi: Implementation gaps and challenges for timely alert. PLoS ONE 2018, 13, e0200858. [Google Scholar] [CrossRef]
- Carrington, C.V.; Auguste, A.J. Evolutionary and ecological factors underlying the tempo and distribution of yellow fever virus activity. Infect. Genet. Evol. 2013, 13, 198–210. [Google Scholar] [CrossRef] [PubMed]
- von Lindern, J.J.; Aroner, S.; Barrett, N.D.; Wicker, J.A.; Davis, C.T.; Barrett, A.D.T. Genome analysis and phylogenetic relationships between east, central and west African isolates of Yellow fever virus. J. Gen. Virol. 2006, 87 Pt 4, 895–907. [Google Scholar] [CrossRef]
- Lee, E.; Lobigs, M. E protein domain III determinants of yellow fever virus 17D vaccine strain enhance binding to glycosaminoglycans, impede virus spread, and attenuate virulence. J. Virol. 2008, 82, 6024–6033. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, M.C.; Gómez, M.M.; Dos Santos, A.A.; Abreu, F.V.S.; Ferreira-de-Brito, A.; Miranda, R.M.; Castro, M.G.; Lourenço-de-Oliveira, R. Genome analysis of yellow fever virus of the ongoing outbreak in Brazil reveals polymorphisms. Mem. Inst. Oswaldo Cruz. 2017, 112, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D.; Higgs, S. Yellow fever: A disease that has yet to be conquered. Annu. Rev. Entomol. 2007, 52, 209–229. [Google Scholar] [CrossRef] [PubMed]
- Schlagenhauf, P.; Chen, L.H. Yellow Fever importation to China—A failure of pre- and post-travel control systems? Int. J. Infect. Dis. 2017, 60, 91–92. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.V.; Murad, S.D.; van der Eijk, A.A.; Metselaar, H.J.; Hartog, H.; Harinck, F.; GeurtsvanKessel, C.H.; Molenkamp, R.; Cotten, M.; Koopmans, M.P. Genomic sequence of yellow fever virus from a Dutch traveller returning from the Gambia-Senegal region, the Netherlands, November 2018. Eur. Surveill. 2019, 24, 1560–7917. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.E.; Holmes, E.C.; Barrett, A.D. Out of Africa: A molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog. 2007, 3, 0030075. [Google Scholar] [CrossRef] [PubMed]
- Kamgang, B.; Vazeille, M.; Yougang, A.P.; Tedjou, A.N.; Wilson-Bahun, T.A.; Mousson, L.; Wondji, C.S.; Failloux, A.B. Potential of Aedes albopictus and Aedes aegypti (Diptera: Culicidae) to transmit yellow fever virus in urban areas in Central Africa. Emerg. Microbe. Infect. 2019, 8, 1636–1641. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
SIMO TCHETGNA, H.; DESCORPS-DECLERE, S.; SELEKON, B.; GARBA-OUANGOLE, S.; KONAMNA, X.; SOUNGOUZA, M.; TEKPA, G.; SOMSE, P.; NAKOUNE, E.; BERTHET, N. Continuous Circulation of Yellow Fever among Rural Populations in the Central African Republic. Viruses 2022, 14, 2014. https://doi.org/10.3390/v14092014
SIMO TCHETGNA H, DESCORPS-DECLERE S, SELEKON B, GARBA-OUANGOLE S, KONAMNA X, SOUNGOUZA M, TEKPA G, SOMSE P, NAKOUNE E, BERTHET N. Continuous Circulation of Yellow Fever among Rural Populations in the Central African Republic. Viruses. 2022; 14(9):2014. https://doi.org/10.3390/v14092014
Chicago/Turabian StyleSIMO TCHETGNA, Huguette, Stéphane DESCORPS-DECLERE, Benjamin SELEKON, Sandra GARBA-OUANGOLE, Xavier KONAMNA, Mathieu SOUNGOUZA, Gaspard TEKPA, Pierre SOMSE, Emmanuel NAKOUNE, and Nicolas BERTHET. 2022. "Continuous Circulation of Yellow Fever among Rural Populations in the Central African Republic" Viruses 14, no. 9: 2014. https://doi.org/10.3390/v14092014




