Equus caballus Papillomavirus Type-9 (EcPV9): First Detection in Asymptomatic Italian Horses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genital Brush Samples Collection from Horses for EcPV9 Detection
2.2. DNA Extraction and Real-Time PCR Targeting EcPV9-L1 and EcPV2-L1
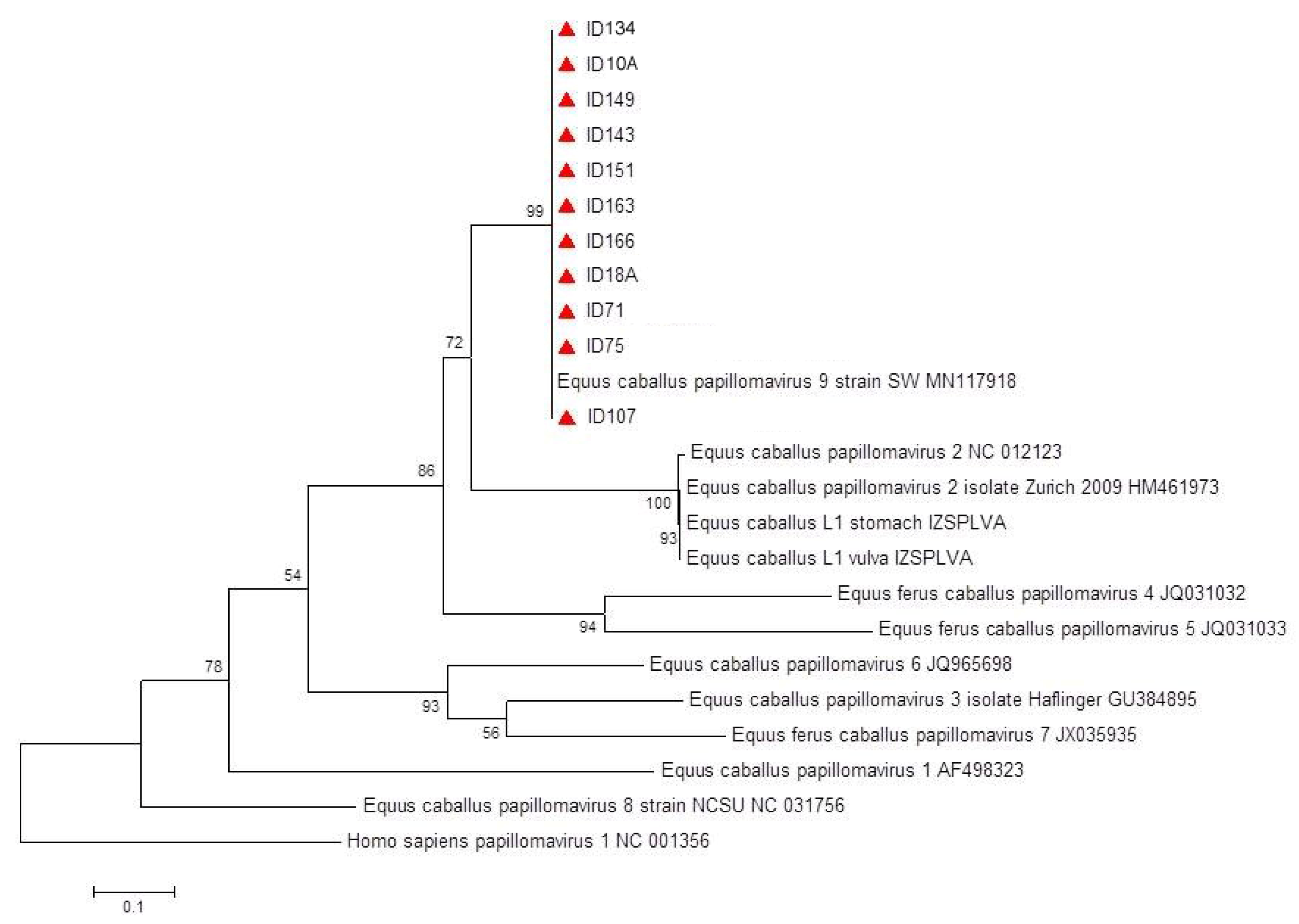
2.3. L1 Sequencing Results and Molecular/Phylogenetic Analysis
| Gene | Sequences | Amplicon Length | Accession | Reference |
|---|---|---|---|---|
| EcPV9-L1 | F-5′- TTC ATC CCA GCT TGA GAC CA-3′ | 116 | MN117918.1 | This paper |
| R-5′-GCA GAT CAA TGG TCC AGA AGG -3′ | ||||
| EcPV9-L1 seq | F-5′-AGG AGA TGT ATG TTG CCC GT -3′ | 533 | ||
| p-EcPV9-L1 | p-FAM- ATT GCC TCC TCA GCC ACC CG-TAMRA | |||
| EcPV2-L1 | F-5′-TTGTCCAGGAGAGGGGTTAG-3′ | 80 | NC_012123 | [26] |
| R-5′-TGCCTTCCTTTTCTTGGTGG-3′ | ||||
| p-EcPV2-L1 | p-FAM-CGTCCAGCACCTTCGACCACCA-TAMRA | |||
| B2M | F-5′-CTGATGTTCTCCAGGTGTTCC-3′ | 114 | NM_001082502.3 | [26] |
| R-5′-TCAATCTCAGGCGGATGGAA-3′ | ||||
| p-B2M | p-FAM-ACTCACGTCACCCAGCAGAGA-TAMRA |
2.4. NGS and Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
3.1. Sampled Horses
3.2. Detection of EcPV9-L1 and EcPV2-L1
3.3. Detection of EcPV9 in Mares
3.4. L1 Sequencing Results and Molecular/Phylogenetic Analysis
3.5. NGS and Bioinformatic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doorbar, J. Host Control of Human Papillomavirus Infection and Disease. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30 (Suppl. 5), F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, S.; Chow-Lockerbie, B.; Ramsauer, S.; Wachoski-Dark, G.; Knight, C.; Wobeser, B. Prevalence of Equus Caballus Papillomavirus Type-2 Infection and Seropositivity in Asymptomatic Western Canadian Horses. Vet. Pathol. 2020, 57, 632–641. [Google Scholar] [CrossRef]
- Araldi, R.P.; Assaf, S.M.R.; Carvalho, R.F.D.; Carvalho, M.A.C.R.D.; Souza, J.M.D.; Magnelli, R.F.; Módolo, D.G.; Roperto, F.P.; Stocco, R.D.C.; Beçak, W. Papillomaviruses: A Systematic Review. Genet. Mol. Biol. 2017, 40, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Yamashita-Kawanishi, N.; Haga, T. Anogenital-Associated Papillomaviruses in Animals: Focusing on Bos Taurus Papillomaviruses. Pathogens 2020, 9, 993. [Google Scholar] [CrossRef] [PubMed]
- Munday, J.S.; Thomson, N.A.; Luff, J.A. Papillomaviruses in Dogs and Cats. Vet. J. 2017, 225, 23–31. [Google Scholar] [CrossRef]
- Cherif, S.; Amine, A.; Thies, S.; Taube, E.T.; Braicu, E.I.; Sehouli, J.; Kaufmann, A.M. Prevalence of Human Papillomavirus Detection in Ovarian Cancer: A Meta-Analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1791–1802. [Google Scholar] [CrossRef]
- Schiffman, M.; Castle, P.E.; Jeronimo, J.; Rodriguez, A.C.; Wacholder, S. Human Papillomavirus and Cervical Cancer. Lancet 2007, 370, 890–907. [Google Scholar] [CrossRef]
- Carter, J.R.; Ding, Z.; Rose, B.R. HPV Infection and Cervical Disease: A Review. Aust. N. Z. J. Obstet. Gynaecol. 2011, 51, 103–108. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primer 2020, 6, 92. [Google Scholar] [CrossRef]
- Guan, P.; Howell-Jones, R.; Li, N.; Bruni, L.; de Sanjosé, S.; Franceschi, S.; Clifford, G.M. Human Papillomavirus Types in 115,789 HPV-Positive Women: A Meta-Analysis from Cervical Infection to Cancer. Int. J. Cancer 2012, 131, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Franceschi, S.; Howell-Jones, R.; Snijders, P.J.F.; Clifford, G.M. Human Papillomavirus Type Distribution in 30,848 Invasive Cervical Cancers Worldwide: Variation by Geographical Region, Histological Type and Year of Publication. Int. J. Cancer 2011, 128, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Ramsauer, A.S.; Wachoski-Dark, G.L.; Fraefel, C.; Tobler, K.; Brandt, S.; Knight, C.G.; Favrot, C.; Grest, P. Paving the Way for More Precise Diagnosis of EcPV2-Associated Equine Penile Lesions. BMC Vet. Res. 2019, 15, 356. [Google Scholar] [CrossRef]
- Bergin, I.L.; Bell, J.D.; Chen, Z.; Zochowski, M.K.; Chai, D.; Schmidt, K.; Culmer, D.L.; Aronoff, D.M.; Patton, D.L.; Mwenda, J.M.; et al. Novel Genital Alphapapillomaviruses in Baboons (Papio Hamadryas Anubis) with Cervical Dysplasia. Vet. Pathol. 2013, 50, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Gottschling, M.; Bravo, I.G.; Schulz, E.; Bracho, M.A.; Deaville, R.; Jepson, P.D.; Van Bressem, M.-F.; Stockfleth, E.; Nindl, I. Modular Organizations of Novel Cetacean Papillomaviruses. Mol. Phylogenet. Evol. 2011, 59, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Nafz, J.; Schäfer, K.; Chen, S.F.; Bravo, I.G.; Ibberson, M.; Nindl, I.; Stockfleth, E.; Rösl, F. A novel rodent papillomavirus isolated from anogenital lesions in its natural host. Virology 2008, 374, 186–197. [Google Scholar] [CrossRef]
- Ingle, A.; Ghim, S.; Joh, J.; Chepkoech, I.; Jenson, A.B.; Sundberg, J.P. Novel Laboratory Mouse Papillomavirus (MusPV) Infection. Vet. Pathol. 2011, 48, 500–505. [Google Scholar] [CrossRef]
- Wu, Z.; Ren, X.; Yang, L.; Hu, Y.; Yang, J.; He, G.; Zhang, J.; Dong, J.; Sun, L.; Du, J.; et al. Virome Analysis for Identification of Novel Mammalian Viruses in Bat Species from Chinese Provinces. J. Virol. 2012, 86, 10999–11012. [Google Scholar] [CrossRef]
- Rojas-Anaya, E.; Cantú-Covarrubias, A.; Álvarez, J.F.M.; Loza-Rubio, E. Detection and Phylogenetic Analysis of Bovine Papillomavirus in Cutaneous Warts in Cattle in Tamaulipas, Mexico. Can. J. Vet. Res. 2016, 80, 262–268. [Google Scholar]
- Li, C.-X.; Chang, W.-S.; Mitsakos, K.; Rodger, J.; Holmes, E.C.; Hudson, B.J. Identification of a Novel Equine Papillomavirus in Semen from a Thoroughbred Stallion with a Penile Lesion. Viruses 2019, 11, 713. [Google Scholar] [CrossRef]
- Sykora, S.; Samek, L.; Schönthaler, K.; Palm, F.; Borzacchiello, G.; Aurich, C.; Brandt, S. EcPV-2 is Transcriptionally Active in Equine SCC but Only Rarely Detectable in Swabs and Semen from Healthy Horses. Vet. Microbiol. 2012, 158, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Sykora, S.; Brandt, S. Papillomavirus Infection and Squamous Cell Carcinoma in Horses. Vet. J. 2017, 223, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Peters-Kennedy, J.; Lange, C.E.; Rine, S.L.; Hackett, R.P. Equus Caballus Papillomavirus 8 (EcPV8) Associated with Multiple Viral Plaques, Viral Papillomas, and Squamous Cell Carcinoma in a Horse. Equine Vet. J. 2019, 51, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and High-Performance Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Mecocci, S.; Porcellato, I.; Armando, F.; Mechelli, L.; Brachelente, C.; Pepe, M.; Gialletti, R.; Passeri, B.; Modesto, P.; Ghelardi, A.; et al. Equine Genital Squamous Cell Carcinoma Associated with EcPV2 Infection: RANKL Pathway Correlated to Inflammation and Wnt Signaling Activation. Biology 2021, 10, 244. [Google Scholar] [CrossRef]
- Andrews, S.; FASTQC. A Quality Control Tool for High Throughput Sequence Data | BibSonomy. Available online: https://www.bibsonomy.org/bibtex/f230a919c34360709aa298734d63dca3 (accessed on 5 August 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tcherepanov, V.; Ehlers, A.; Upton, C. Genome Annotation Transfer Utility (GATU): Rapid Annotation of Viral Genomes Using a Closely Related Reference Genome. BMC Genom. 2006, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Porcellato, I.; Modesto, P.; Cappelli, K.; Varello, K.; Peletto, S.; Brachelente, C.; Martini, I.; Mechelli, L.; Ferrari, A.; Ghelardi, A.; et al. Equus Caballus Papillomavirus Type 2 (EcPV2) in Co-Occurring Vulvar and Gastric Lesions of a Pony. Res. Vet. Sci. 2020, 132, 167–171. [Google Scholar] [CrossRef]
- Cappelli, K.; Ciucis, C.G.D.; Mecocci, S.; Nervo, T.; Crescio, M.I.; Pepe, M.; Gialletti, R.; Pietrucci, D.; Migone, L.F.; Turco, S.; et al. Detection of Equus Caballus Papillomavirus Type-2 in Asymptomatic Italian Horses. Viruses 2022, 14, 1696. [Google Scholar] [CrossRef]
- Aguilar, P.D.; González, C.L.; Rodríguez, A.C.; Páez, K.A.; Arévalo, A.P.; Bobokova, J. Prevalence of High-Risk Genotypes of Human Papillomavirus: Women Diagnosed with Premalignant and Malignant Pap Smear Tests in Southern Ecuador. Infect. Dis. Obstet. Gynecol. 2017, 2017, e8572065. [Google Scholar] [CrossRef]
- Bosch, F.X.; Lorincz, A.; Muñoz, N.; Meijer, C.J.L.M.; Shah, K.V. The Causal Relation between Human Papillomavirus and Cervical Cancer. J. Clin. Pathol. 2002, 55, 244–265. [Google Scholar] [CrossRef]
- Mira, J.; Herman, M.; Zakia, L.S.; Olivo, G.; Araújo, J.P.; Borges, A.S.; Oliveira-Filho, J.P. Frequency of Equus Caballus Papillomavirus in Equine Aural Plaques. J. Vet. Diagn. Investig. 2018, 30, 565–568. [Google Scholar] [CrossRef]
- Lange, C.E.; Vetsch, E.; Ackermann, M.; Favrot, C.; Tobler, K. Four Novel Papillomavirus Sequences Support a Broad Diversity among Equine Papillomaviruses. J. Gen. Virol. 2013, 94, 1365–1372. [Google Scholar] [CrossRef]
- Dong, J.; Zhu, W.; Yamashita, N.; Chambers, J.K.; Uchida, K.; Kuwano, A.; Haga, T. Isolation of Equine Papillomavirus Type 1 from Racing Horse in Japan. J. Vet. Med. Sci. 2017, 79, 1957–1959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, K.E.; Bizikova, P.; Luff, J.; Zhou, D.; Yuan, H.; Breuhaus, B.; Nelson, E.; Mackay, R. Generalized Papillomatosis in Three Horses Associated with a Novel Equine Papillomavirus (EcPV8). Vet. Dermatol. 2018, 29, 72-e30. [Google Scholar] [CrossRef] [PubMed]
- Hibi, H.; Hatama, S.; Obata, A.; Shibahara, T.; Kadota, K. Laryngeal Squamous Cell Carcinoma and Papilloma Associated with Equus Caballus Papillomavirus 2 in a Horse. J. Vet. Med. Sci. 2019, 81, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Van den Top, J.G.B.; de Heer, N.; Klein, W.R.; Ensink, J.M. Penile and Preputial Squamous Cell Carcinoma in the Horse: A Retrospective Study of Treatment of 77 Affected Horses. Equine Vet. J. 2008, 40, 533–537. [Google Scholar] [CrossRef]
- Scott, D.W.; Miller, W.H. Equine Dermatology. J. Equine Vet. Sci. 2003, 2, 65–67. [Google Scholar] [CrossRef]
- Straticò, P.; Razzuoli, E.; Hattab, J.; Guerri, G.; Celani, G.; Palozzo, A.; Bonanni, D.; Fruscione, F.; Varasano, V.; Petrizzi, L.; et al. Equine Gastric Squamous Cell Carcinoma in a Friesian Stallion. J. Equine Vet. Sci. 2022, 117, 104087. [Google Scholar] [CrossRef]
- Alloway, E.; Linder, K.; May, S.; Rose, T.; DeLay, J.; Bender, S.; Tucker, A.; Luff, J. A Subset of Equine Gastric Squamous Cell Carcinomas is Associated with Equus Caballus Papillomavirus-2 Infection. Vet. Pathol. 2020, 57, 427–431. [Google Scholar] [CrossRef]
- Lange, C.E.; Tobler, K.; Lehner, A.; Grest, P.; Welle, M.M.; Schwarzwald, C.C.; Favrot, C. EcPV2 DNA in Equine Papillomas and In Situ and Invasive Squamous Cell Carcinomas Supports Papillomavirus Etiology. Vet. Pathol. 2013, 50, 686–692. [Google Scholar] [CrossRef]
- Taylor, S.D.; Haldorson, G.J.; Vaughan, B.; Pusterla, N. Gastric Neoplasia in Horses. J. Vet. Intern. Med. 2009, 23, 1097–1102. [Google Scholar] [CrossRef]
- Padilla-Mendoza, J.R.; Gómez-López, L.A.; López-Casamichana, M.; Azuara-Liceaga, E.I.; Cortés-Malagón, E.M.; López-Cánovas, L.; Reyes-Hernández, O.D.; Rodríguez, M.A.; Bonilla-Delgado, J.; López-Reyes, I. Human Papillomavirus Coinfection in the Cervical Intraepithelial Lesions and Cancer of Mexican Patients. BioMed Res. Int. 2020, 2020, 4542320. [Google Scholar] [CrossRef]
- Guo, Z.; Hu, Y.; Yuan, L.; Li, N.; Wang, T. A Prospective Study on the Predictive Value of DNA Methylation in Cervical Intraepithelial Neoplasia Prognosis. Arch. Gynecol. Obstet. 2018, 298, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Bello, J.O.; Carrillo-García, A.; Lizano, M. Epidemiology and Molecular Biology of HPV Variants in Cervical Cancer: The State of the Art in Mexico. Int. J. Mol. Sci. 2022, 23, 8566. [Google Scholar] [CrossRef]
- Zaugg, N.; Nespeca, G.; Hauser, B.; Ackermann, M.; Favrot, C. Detection of Novel Papillomaviruses in Canine Mucosal, Cutaneous and in Situ Squamous Cell Carcinomas. Vet. Dermatol. 2005, 16, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ghim, S.; Newsome, J.; Apolinario, T.; Olcese, V.; Martin, M.; Delius, H.; Felsburg, P.; Jenson, B.; Schlegel, R. An Epidermotropic Canine Papillomavirus with Malignant Potential Contains an E5 Gene and Establishes a Unique Genus. Virology 2007, 359, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.E.; Tobler, K.; Markau, T.; Alhaidari, Z.; Bornand, V.; Stöckli, R.; Trüssel, M.; Ackermann, M.; Favrot, C. Sequence and Classification of FdPV2, a Papillomavirus Isolated from Feline Bowenoid in Situ Carcinomas. Vet. Microbiol. 2009, 137, 60–65. [Google Scholar] [CrossRef]
- Vitiello, V.; Burrai, G.P.; Pisanu, S.; Cacciotto, C.; Addis, M.F.; Alberti, A.; Antuofermo, E.; Cubeddu, T.; Pirino, S. Proteomic Profiles and Cytokeratin 13 as a Potential Biomarker of Ovis Aries Papillomavirus 3-Positive and Negative Cutaneous Squamous Cell Carcinomas. Res. Vet. Sci. 2021, 134, 112–119. [Google Scholar] [CrossRef]
- Vitiello, V.; Burrai, G.P.; Agus, M.; Anfossi, A.G.; Alberti, A.; Antuofermo, E.; Rocca, S.; Cubeddu, T.; Pirino, S. Ovis Aries Papillomavirus 3 in Ovine Cutaneous Squamous Cell Carcinoma. Vet. Pathol. 2017, 54, 775–782. [Google Scholar] [CrossRef]
- Zur Hausen, H. Papillomaviruses Causing Cancer: Evasion from Host-Cell Control in Early Events in Carcinogenesis. JNCI J. Natl. Cancer Inst. 2000, 92, 690–698. [Google Scholar] [CrossRef]
- Zur Hausen, H. Papillomavirus Infections—A Major Cause of Human Cancers. Biochim. Biophys. Acta BBA Rev. Cancer 1996, 1288, F55–F78. [Google Scholar] [CrossRef]
- Dayyani, F.; Etzel, C.J.; Liu, M.; Ho, C.-H.; Lippman, S.M.; Tsao, A.S. Meta-Analysis of the Impact of Human Papillomavirus (HPV) on Cancer Risk and Overall Survival in Head and Neck Squamous Cell Carcinomas (HNSCC). Head Neck Oncol. 2010, 2, 15. [Google Scholar] [CrossRef]
- Tovar, J.M.; Bazaldua, O.V.; Vargas, L.; Reile, E. Human Papillomavirus, Cervical Cancer, and the Vaccines. Postgrad. Med. 2008, 120, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Kombe, A.J.K.; Li, B.; Zahid, A.; Mengist, H.M.; Bounda, G.-A.; Zhou, Y.; Jin, T. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front. Public Health 2021, 8, 552028. [Google Scholar] [CrossRef] [PubMed]
| EcPV Type | Positive |
|---|---|
| Equus caballus papillomavirus type 1 | AF498323.1 |
| Equus caballus papillomavirus type 1 | NC_003748.1 |
| Equus caballus papillomavirus type 1 strain 150904 | MF288893.1 |
| Equus caballus papillomavirus type 1 isolate G2 | MN164462.1 |
| Equine papillomavirus 2 | EU503122.1 |
| Equine papillomavirus 2 | NC_012123.1 |
| Equine papillomavirus 2 isolate Zurich 2009 | HM461973.1 |
| Equus caballus papillomavirus 2 isolate XJ-ks1391 | MW410986.1 |
| Equine papillomavirus 3 | NC_017862.1 |
| Equine papillomavirus 3 isolate Haflinger | GU384895.1 |
| Equus ferus caballus papillomavirus type 4 | NC_020085.1 |
| Equus ferus caballus papillomavirus type 4 | JQ031032.1 |
| Equus ferus caballus papillomavirus type 5 | JQ031033.1 |
| Equus ferus caballus papillomavirus type 5 | NC_020084.1 |
| Equine papillomavirus type 6 | JQ965698.1 |
| Equine papillomavirus type 6 | NC_020500.1 |
| Equus ferus caballus papillomavirus type 7 | JX035935.1 |
| Equus ferus caballus papillomavirus type 7 | NC_020501.1 |
| Equus caballus papillomavirus type 8 strain NCSU | KU963288.1 |
| Equus caballus papillomavirus type 8 strain NCSU | NC_031756.1 |
| Equus caballus papillomavirus 9 strain SW | MN117918.1 |
| Positive | % | Negative | % | Total | |
|---|---|---|---|---|---|
| English Thoroughbred | 10 | 13.9% | 62 | 86.1% | 72 |
| Shire | 1 | 25% | 3 | 75% | 4 |
| <6 yy | 6–<9 yy | 9–<13 yy | ≥13 yy | |
|---|---|---|---|---|
| Positive | 0 | 2 | 6 | 1 |
| Negative | 41 | 32 | 56 | 63 |
| Total | 41 | 34 | 62 | 64 |
| Positive | % | Negative | % | Total | |
|---|---|---|---|---|---|
| English Thoroughbred | 10 | 16.1 | 52 | 83.9 | 62 |
| Shire | 1 | 25.0 | 3 | 75.0 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Paolis, L.; De Ciucis, C.G.; Peletto, S.; Cappelli, K.; Mecocci, S.; Nervo, T.; Guardone, L.; Crescio, M.I.; Pietrucci, D.; Fruscione, F.; et al. Equus caballus Papillomavirus Type-9 (EcPV9): First Detection in Asymptomatic Italian Horses. Viruses 2022, 14, 2050. https://doi.org/10.3390/v14092050
De Paolis L, De Ciucis CG, Peletto S, Cappelli K, Mecocci S, Nervo T, Guardone L, Crescio MI, Pietrucci D, Fruscione F, et al. Equus caballus Papillomavirus Type-9 (EcPV9): First Detection in Asymptomatic Italian Horses. Viruses. 2022; 14(9):2050. https://doi.org/10.3390/v14092050
Chicago/Turabian StyleDe Paolis, Livia, Chiara Grazia De Ciucis, Simone Peletto, Katia Cappelli, Samanta Mecocci, Tiziana Nervo, Lisa Guardone, Maria Ines Crescio, Daniele Pietrucci, Floriana Fruscione, and et al. 2022. "Equus caballus Papillomavirus Type-9 (EcPV9): First Detection in Asymptomatic Italian Horses" Viruses 14, no. 9: 2050. https://doi.org/10.3390/v14092050
APA StyleDe Paolis, L., De Ciucis, C. G., Peletto, S., Cappelli, K., Mecocci, S., Nervo, T., Guardone, L., Crescio, M. I., Pietrucci, D., Fruscione, F., Gabbianelli, F., Turco, S., Varello, K., Donato, G. G., Maurella, C., Modesto, P., Maniaci, M. G., Chillemi, G., Ghelardi, A., & Razzuoli, E. (2022). Equus caballus Papillomavirus Type-9 (EcPV9): First Detection in Asymptomatic Italian Horses. Viruses, 14(9), 2050. https://doi.org/10.3390/v14092050








