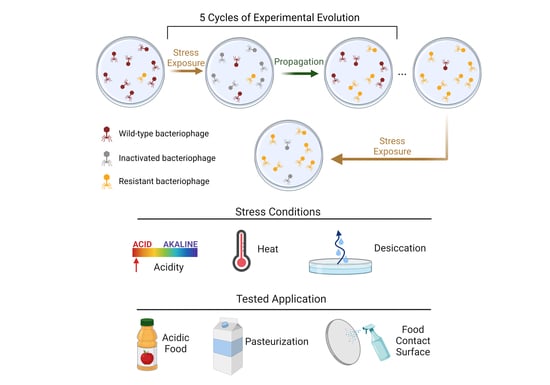
Stress Exposure of Evolved Bacteriophages under Laboratory versus Food Processing Conditions Highlights Challenges in Translatability
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria and Bacteriophage Strains
2.2. Chemicals
2.3. Bacteria Culture, Bacteriophage Propagation, and Quantification
2.4. Bacteriophage Selection under Stress and Reversion-Testing
2.4.1. Vacuum Desiccation
2.4.2. Elevated Temperature
2.4.3. Low pH
2.5. Bacteria DNA Extraction and Sequencing
2.6. Bacteriophage DNA Extraction and Sequencing
2.7. Quantification of Bacteriophage Infectivity
2.7.1. Efficiency of Plaquing and Plaque Morphology
2.7.2. Kill Curves
2.8. Comparison of Application Performance
2.8.1. Bacteriophage Survival on Stainless Steel Surfaces
2.8.2. Bacteriophage Survival within Milk after Batch Pasteurization
2.8.3. Bacteriophage Survival in Apple Juice
2.9. Statistical Analysis
3. Results and Discussion
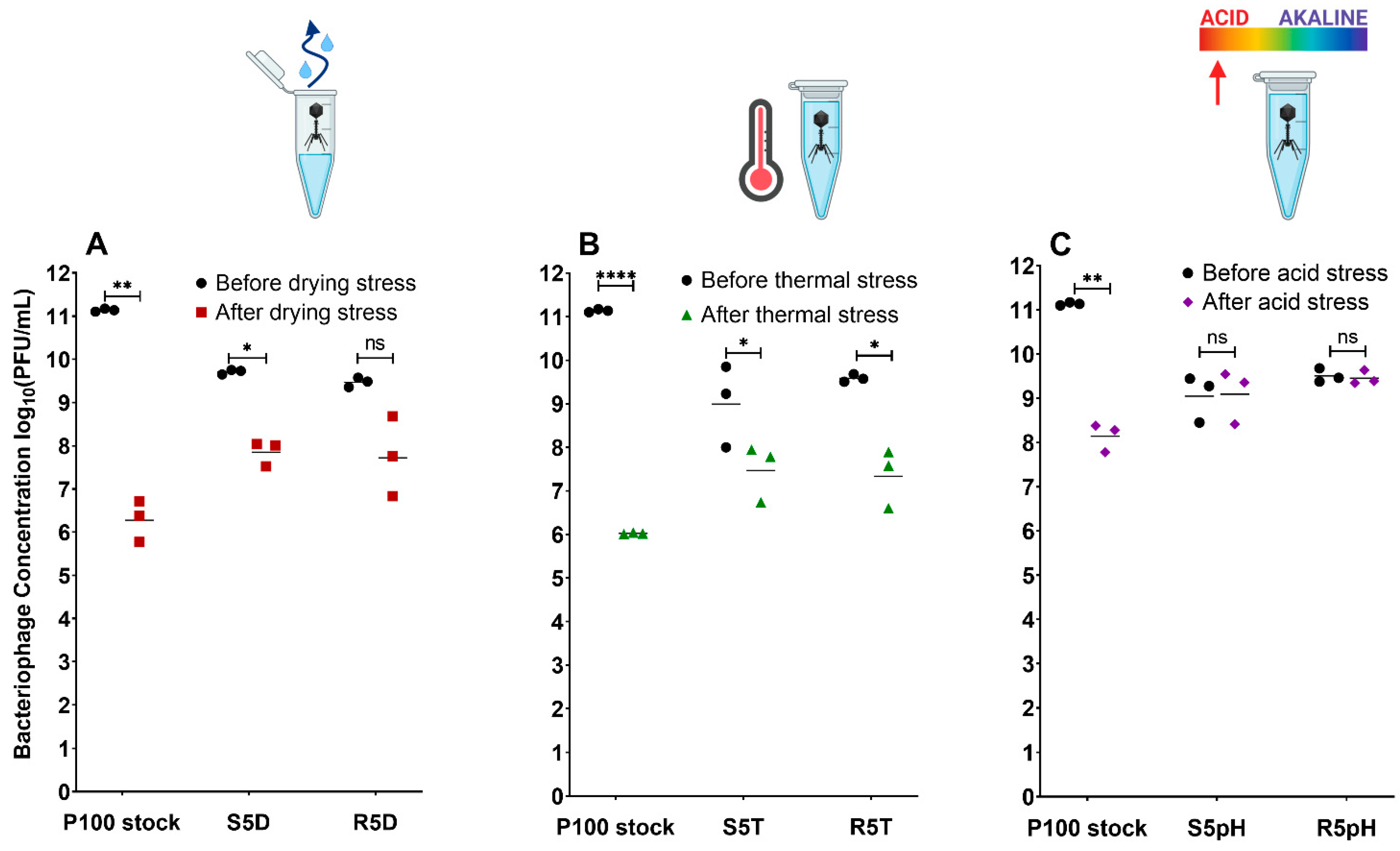
3.1. Enhanced Stress Resistance and Low Reversion Rates for Selected Bacteriophage
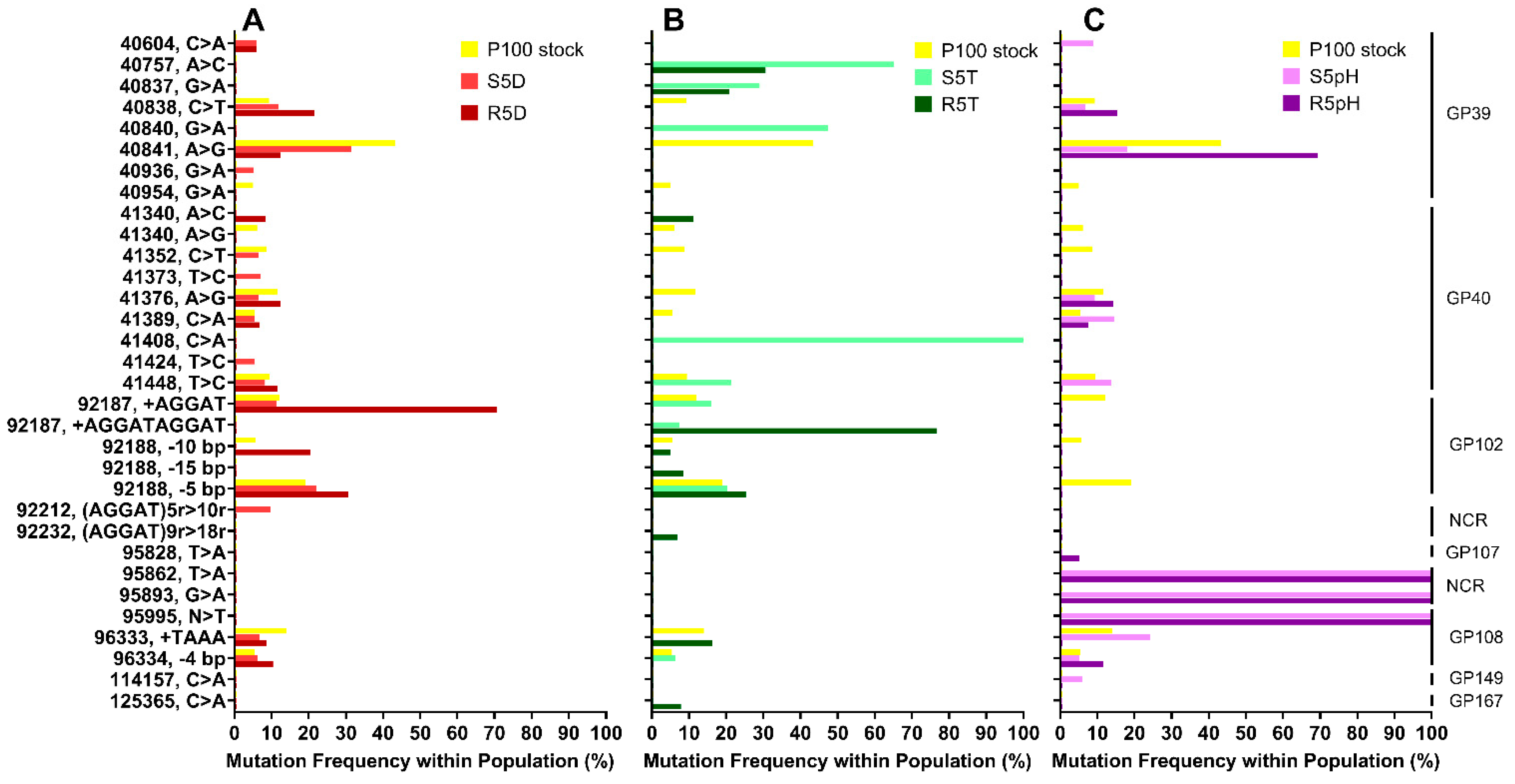
3.2. Genomic Sequences of Stress-Selected Samples and Reversion-Tested Samples
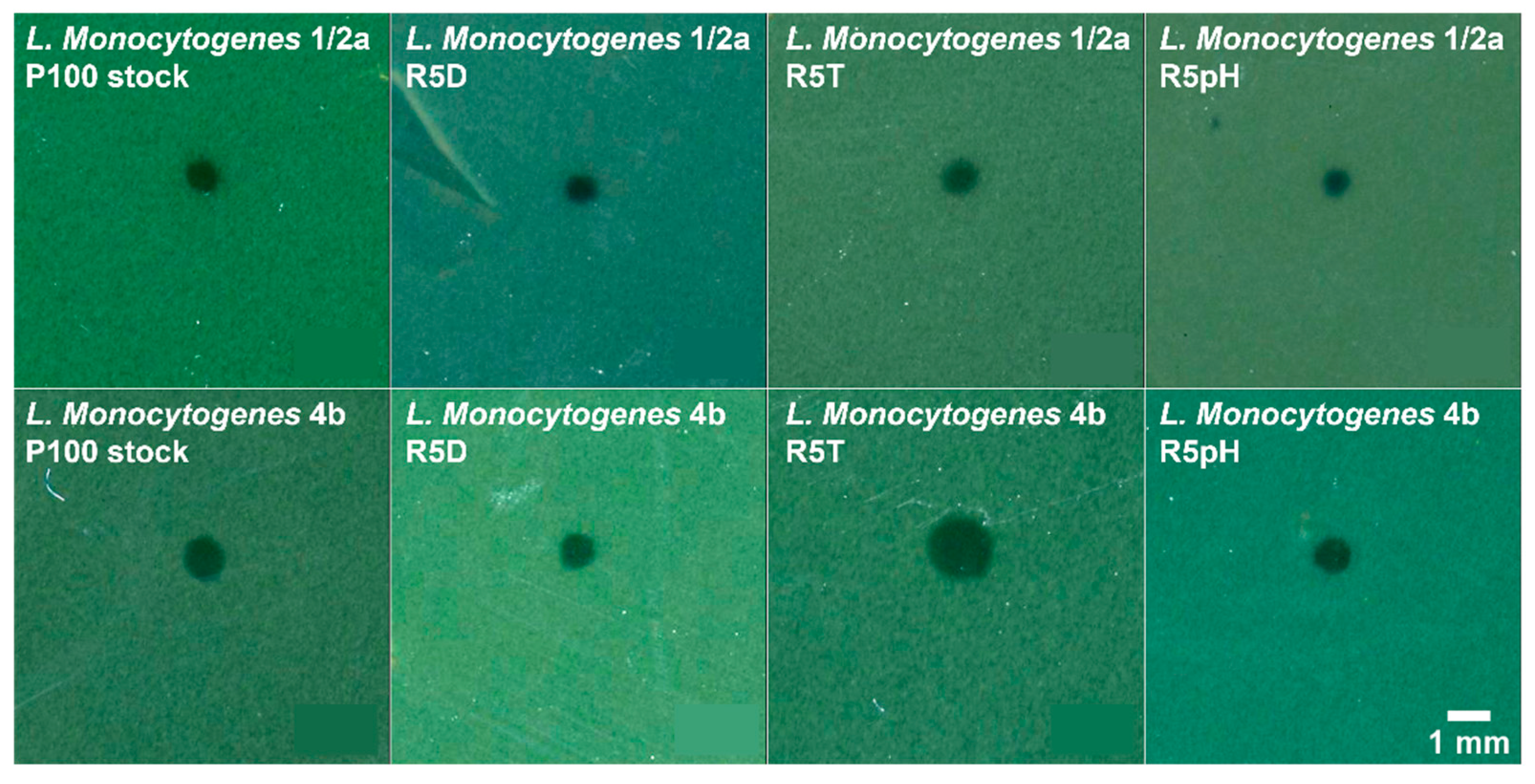
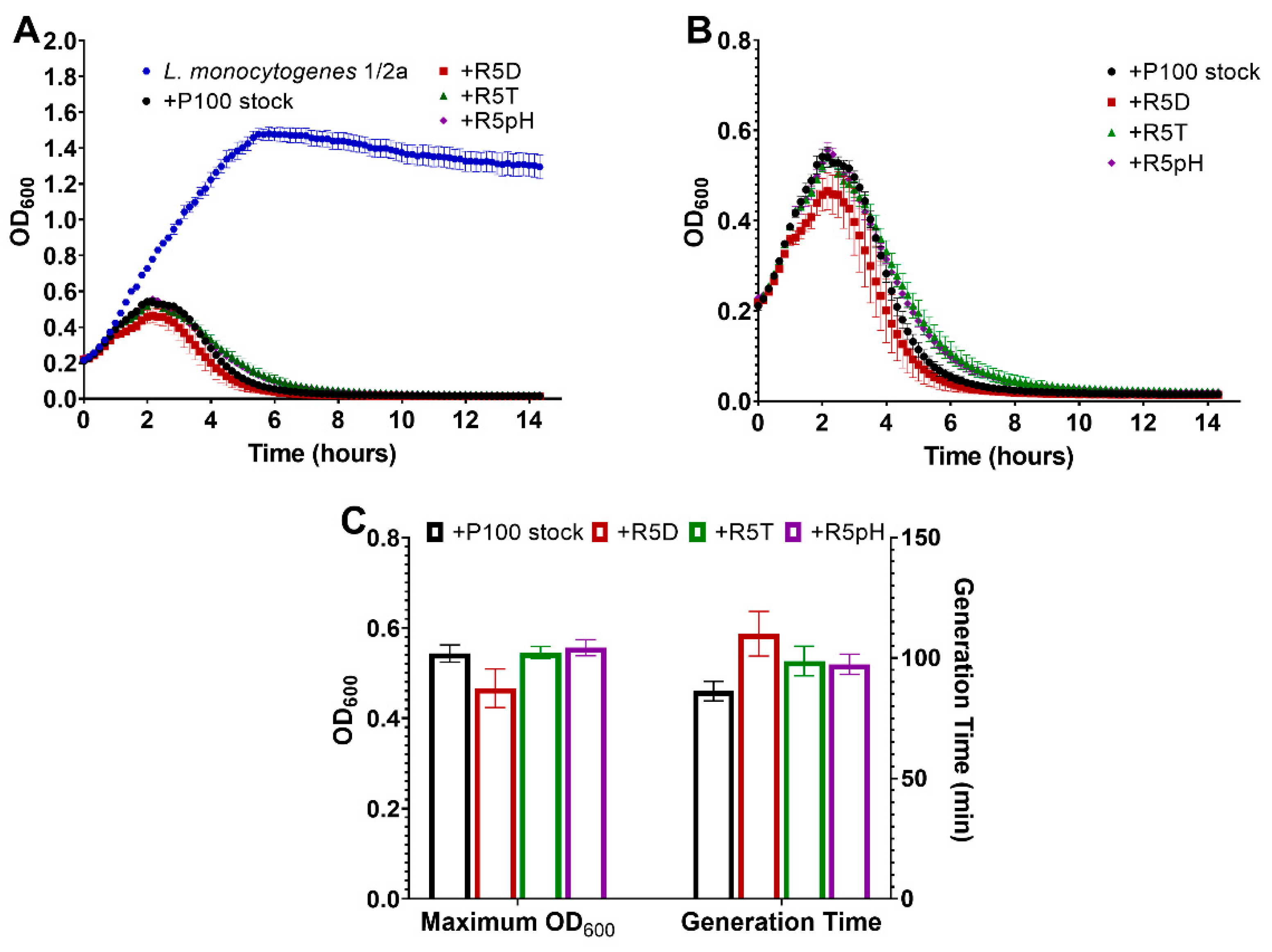
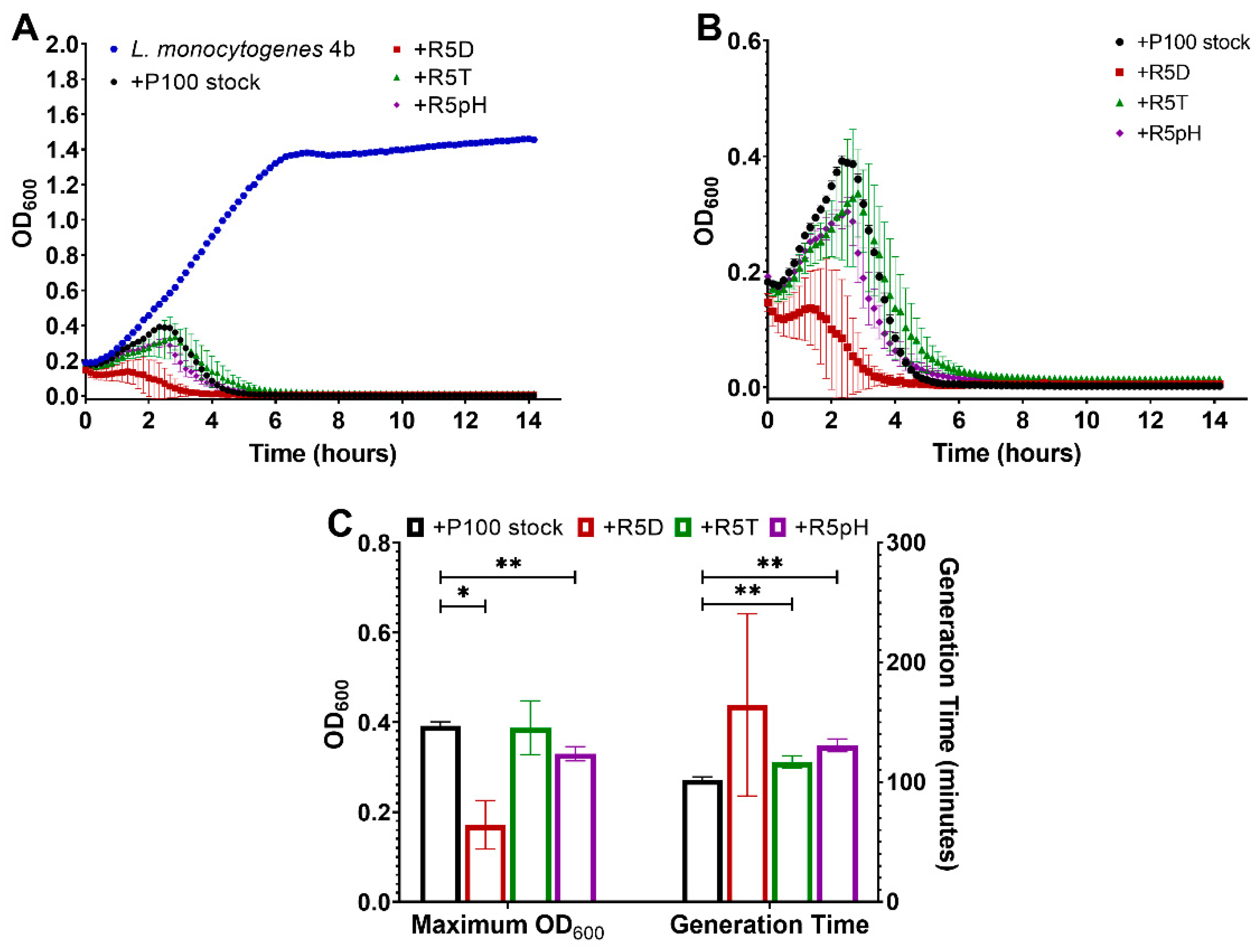
3.3. Antibacterial Potency Preserved in Reversion-Tested Bacteriophage
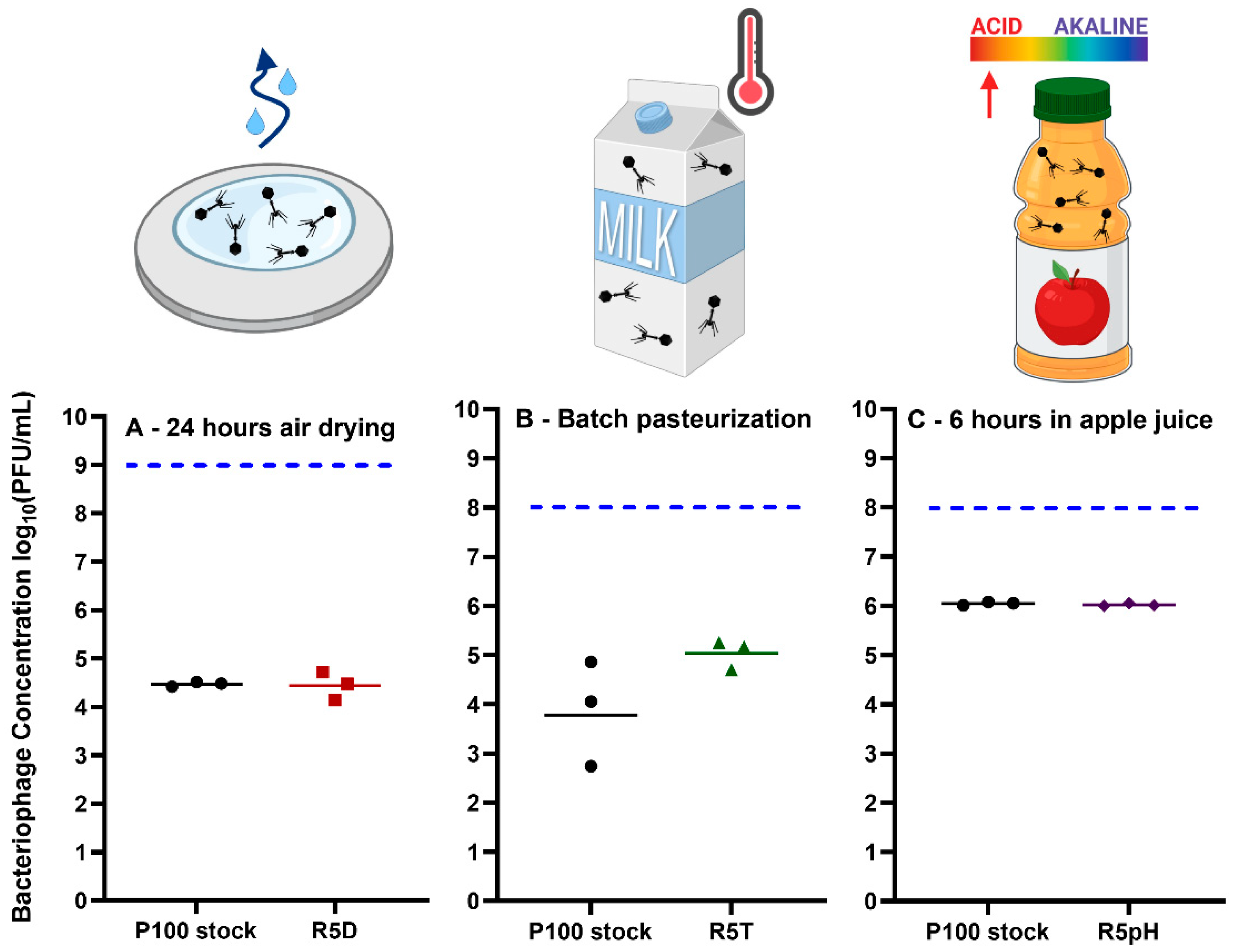
3.4. Comparison of Infectivity for P100 Stock and Reversion-Tested Bacteriophages on Food Preparation Surfaces and in Food Matrices
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nyarko, E.B.; Donnelly, C.W. Listeria monocytogenes: Strain Heterogeneity, Methods, and Challenges of Subtyping. J. Food Sci. 2015, 80, M2868–M2878. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Milian, A.; Payeras-Cifre, A. What Is New in Listeriosis? BioMed Res. Int. 2014, 2014, 358051. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Gooneratne, S.R.; Hussain, M.A. Listeria monocytogenes in fresh produce: Outbreaks, prevalence and contamination levels. Foods 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Gorris, L.G.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- NicAogáin, K.; O’Byrne, C.P. The Role of Stress and Stress Adaptations in Determining the Fate of the Bacterial Pathogen Listeria monocytogenes in the Food Chain. Front. Microbiol. 2016, 7, 1865. [Google Scholar] [CrossRef]
- Faleiro, M.L.; Andrew, P.; Power, D. Stress response of Listeria monocytogenes isolated from cheese and other foods. Int. J. Food Microbiol. 2003, 84, 207–216. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Takahashi, H.; Kuramoto, S.; Miya, S.; Kimura, B. Desiccation survival of Listeria monocytogenes and other potential foodborne pathogens on stainless steel surfaces is affected by different food soils. Food Control 2011, 22, 633–637. [Google Scholar] [CrossRef]
- Zoz, F.; Grandvalet, C.; Lang, E.; Iaconelli, C.; Gervais, P.; Firmesse, O.; Guyot, S.; Beney, L. Listeria monocytogenes ability to survive desiccation: Influence of serotype, origin, virulence, and genotype. Int. J. Food Microbiol. 2017, 248, 82–89. [Google Scholar] [CrossRef]
- Chan, Y.C.; Wiedmann, M. Physiology and Genetics of Listeria Monocytogenes Survival and Growth at Cold Temperatures. Crit. Rev. Food Sci. Nutr. 2008, 49, 237–253. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Reinhard, R.G.; Kalinowski, R.M.; Bodnaruk, P.W.; Eifert, J.D.; Boyer, R.R.; Duncan, S.E.; Bailey, R.H. Practical application of bacteriophage in food manufacturing facilities for the control of Listeria sp. J. Food Saf. 2020, e12871. [Google Scholar] [CrossRef]
- Forauer, E.; Wu, S.T.; Etter, A.J. Listeria monocytogenes in the retail deli environment: A review. Food Control 2021, 119, 107443. [Google Scholar] [CrossRef]
- Endersen, L.; Coffey, A. The use of bacteriophages for food safety. Curr. Opin. Food Sci. 2020, 36, 1–8. [Google Scholar] [CrossRef]
- Łobocka, M.; Dąbrowska, K.; Górski, A. Engineered Bacteriophage Therapeutics: Rationale, Challenges and Future. BioDrugs 2021, 35, 255–280. [Google Scholar] [CrossRef] [PubMed]
- Cristobal-Cueto, P.; García-Quintanilla, A.; Esteban, J.; García-Quintanilla, M. Phages in food industry biocontrol and bioremediation. Antibiotics 2021, 10, 786. [Google Scholar] [CrossRef]
- Fister, S.; Robben, C.; Witte, A.K.; Schoder, D.; Wagner, M.; Rossmanith, P. Influence of Environmental Factors on Phage–Bacteria Interaction and on the Efficacy and Infectivity of Phage P100. Front. Microbiol. 2016, 7, 1152. [Google Scholar] [CrossRef]
- Ahmadi, H.; Radford, D.; Kropinski, A.M.; Lim, L.T.; Balamurugan, S. Thermal-stability and reconstitution ability of Listeria phages P100 and A511. Front. Microbiol. 2017, 8, 2375. [Google Scholar] [CrossRef]
- Leung, V.; Szewczyk, A.; Chau, J.; Hosseinidoust, Z.; Groves, L.; Hawsawi, H.; Anany, H.; Griffiths, M.W.; Ali, M.M.; Filipe, C.D.M. Long-Term Preservation of Bacteriophage Antimicrobials Using Sugar Glasses. ACS Biomater. Sci. Eng. 2017, 4, 3802–3808. [Google Scholar] [CrossRef]
- Leung, V.; Groves, L.; Szewczyk, A.; Hosseinidoust, Z.; Filipe, C.D.M. Long-Term Antimicrobial Activity of Phage-Sugar Glasses is Closely Tied to the Processing Conditions. ACS Omega 2018, 3, 18295–18303. [Google Scholar] [CrossRef]
- Tom, E.F.; Molineux, I.J.; Paff, M.L.; Bull, J.J. Experimental evolution of UV resistance in a phage. PeerJ 2018, 6, e5190. [Google Scholar] [CrossRef] [PubMed]
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses 2020, 12, 423. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, E.; Arribas, M.; Cabanillas, L.; Román, I.; Acosta, E. Evolutionary adaptation of an RNA bacteriophage to the simultaneous increase in the within-host and extracellular temperatures. Sci. Rep. 2018, 8, 8080. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, A.; Sugawara, R.; Tsushima, F.S.; Kumagai, T.; Yomo, T. Contribution of Silent Mutations to Thermal Adaptation of RNA Bacteriophage Qβ. J. Virol. 2014, 88, 11459–11468. [Google Scholar] [CrossRef]
- Perera, M.N.; Abuladze, T.; Li, M.; Woolston, J.; Sulakvelidze, A. Bacteriophage cocktail significantly reduces or eliminates Listeria monocytogenes contamination on lettuce, apples, cheese, smoked salmon and frozen foods. Food Microbiol. 2015, 52, 42–48. [Google Scholar] [CrossRef]
- Vikram, A.; Woolston, J.; Sulakvelidze, A. Phage Biocontrol Applications in Food Production and Processing. Curr. Issues Mol. Biol. 2021, 40, 267–302. [Google Scholar] [CrossRef]
- Carlton, R.M.; Noordman, W.H.; Biswas, B.; de Meester, E.D.; Loessner, M.J. Bacteriophage P100 for control of Listeria monocytogenes in foods: Genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 2005, 43, 301–312. [Google Scholar] [CrossRef]
- Chibeu, A.; Agius, L.; Gao, A.; Sabour, P.M.; Kropinski, A.M.; Balamurugan, S. Efficacy of bacteriophage LISTEXTMP100 combined with chemical antimicrobials in reducing Listeria monocytogenes in cooked turkey and roast beef. Int. J. Food Microbiol. 2013, 167, 208–214. [Google Scholar] [CrossRef]
- Figueiredo, A.C.L.; Almeida, R.C.C. Antibacterial efficacy of nisin, bacteriophage P100 and sodium lactate against Listeria monocytogenes in ready-to-eat sliced pork ham. Braz. J. Microbiol. 2017, 48, 724–729. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Original Articles SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Green, M.R. Molecular Cloning: A Laboratory Manual/Michael R. Green, Joseph Sambrook; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef] [PubMed]
- Carlton, R.M.; Noordman, W.H.; Biswas, B.; de Meester, E.D.; Loessner, M.J. Listeria Bacteriophage P100, Complete Genome—Nucleotide—NCBI. Available online: https://www.ncbi.nlm.nih.gov/nuccore/82547605 (accessed on 11 February 2020).
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Arachchi, G.J.G.; Cridge, A.G.; Dias-Wanigasekera, B.M.; Cruz, C.D.; McIntyre, L.; Liu, R.; Flint, S.H.; Mutukumira, A.N. Effectiveness of phages in the decontamination of Listeria monocytogenes adhered to clean stainless steel, stainless steel coated with fish protein, and as a biofilm. J. Ind. Microbiol. Biotechnol. 2013, 40, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Montañez-Izquierdo, V.Y.; Salas-Vázquez, D.I.; Rodríguez-Jerez, J.J. Use of epifluorescence microscopy to assess the effectiveness of phage P100 in controlling Listeria monocytogenes biofilms on stainless steel surfaces. Food Control 2012, 23, 470–477. [Google Scholar] [CrossRef]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages Using the Small Drop Plaque Assay System. In Methods in Molecular Biology; Clokie, A.M., Martha, J.R., Kropinski, Eds.; Humana Press: Clifton, NJ, USA, 2009; Volume 501, pp. 81–85. [Google Scholar]
- Agency, C.F.I. Dairy Processing: Batch Pasteurization Systems. 2020. Available online: https://inspection.canada.ca/preventive-controls/dairy-products/batch-pasteurization/eng/1539361429523/1539361508211 (accessed on 19 July 2021).
- Micreos Food Safety. PhageGuard Listex Application Data Sheet Cheese. Wageningen, The Netherlands, 2016. Available online: https://www.PhageGuard.com (accessed on 9 May 2022).
- European Food Safety Authority. Evaluation of the safety and efficacy of ListexTM P100 for reduction of pathogens on different ready-to-eat (RTE) food products. EFSA J. 2016, 14, e04565. [Google Scholar] [CrossRef]
- Schaum, C.-E.; Buckling, A.; Smirnoff, N.; Yvon-Durocher, G. Evolution of thermal tolerance and phenotypic plasticity under rapid and slow temperature fluctuations. Proc. R. Soc. B Biol. Sci. 2022, 289, 20220834. [Google Scholar] [CrossRef]
- Peters, T.L.; Song, Y.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Mutant and Recombinant Phages Selected from In Vitro Coevolution Conditions Overcome Phage-Resistant Listeria monocytogenes. Appl. Environ. Microbiol. 2020, 86, e02138-20. [Google Scholar] [CrossRef]
- Bertrand, S.; Ceyssens, P.J.; Yde, M.; Dierick, K.; Boyen, F.; Vanderpas, J.; Vanhoof, R.; Mattheus, W. Diversity of listeria monocytogenes strains of clinical and food chain origins in Belgium between 1985 and 2014. PLoS ONE 2016, 11, e0164283. [Google Scholar] [CrossRef]
- Gianfranceschi, M.V.; D’Ottavio, M.C.; Gattuso, A.; Bella, A.; Aureli, P. Distribution of serotypes and pulsotypes of Listeria monocytogenes from human, food and environmental isolates (Italy 2002–2005). Food Microbiol. 2009, 26, 520–526. [Google Scholar] [CrossRef]
- Shamloo, E.; Hosseini, H.; Moghadam, Z.A.; Larsen, M.H.; Haslberger, A.; Alebouyeh, M. Importance of Listeria monocytogenes in food safety: A review of its prevalence, detection, and antibiotic resistance. Iran. J. Vet. Res. 2019, 20, 241. [Google Scholar] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontéen, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, A.; Kadoya, T.; Kumasaka, N.; Kumagai, T.; Tsushima, F.S.; Yomo, T. Influence of adaptive mutations, from thermal adaptation experiments, on the infection cycle of RNA bacteriophage Qβ. Arch. Virol. 2018, 163, 2655–2662. [Google Scholar] [CrossRef]
- McGee, L.W.; Aitchison, E.W.; Caudle, S.B.; Morrison, A.J.; Zheng, L.; Yang, W.; Rokyta, D.R. Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures. PLoS Genet. 2014, 10, e1004611. [Google Scholar] [CrossRef] [PubMed]
- Micreos Food Safety. PhageGuard Listex Reduces Listeria on Food Contact Surfaces and in the Environment. 2020. Available online: https://phageguard.com/wp-content/uploads/2020/09/FINAL-environmental-application-data-sheet-version-8-SS-SFBdVRAF.pdf (accessed on 2 June 2021).
- Sebastianski, M.; Bridger, N.A.; Featherstone, R.M.; Robinson, J.L. Disease outbreaks linked to pasteurized and unpasteurized dairy products in Canada and the United States: A systematic review. Can. J. Public Health 2022, 113, 569–578. [Google Scholar] [CrossRef] [PubMed]
- USPHS/FDA Grade ‘A’ Pasteurized Milk Ordinance (PMO). U.S. Department of Health and Human Services Public Health Service Food and Drug Administration. 2017. Available online: https://www.fda.gov/downloads/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/Milk/UCM612027.pdf (accessed on 18 July 2022).
- Komora, N.; Bruschi, C.; Ferreira, V.; Maciel, C.; Brandão, T.R.; Fernandes, R.; Saraiva, J.A.; Castro, S.M.; Teixeira, P. The protective effect of food matrices on Listeria lytic bacteriophage P100 application towards high pressure processing. Food Microbiol. 2018, 76, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yamamoto, E.; Murphy, J.; Locas, A. Microbiological safety of ready-to-eat fresh-cut fruits and vegetables sold on the Canadian retail market. Int. J. Food Microbiol. 2020, 335, 108855. [Google Scholar] [CrossRef]
- Oliveira, M.; Viñas, I.; Colàs, P.; Anguera, M.; Usall, J.; Abadias, M. Effectiveness of a bacteriophage in reducing Listeria monocytogenes on fresh-cut fruits and fruit juices. Food Microbiol. 2014, 38, 137–142. [Google Scholar] [CrossRef]
- Leverentz, B.; Conway, W.S.; Janisiewicz, W.; Camp, M.J. Optimizing concentration and timing of a phage spray application to reduce Listeria monocytogenes on honeydew melon tissue. J. Food Prot. 2004, 67, 1682–1686. [Google Scholar] [CrossRef]
- Bumunang, E.W.; Ateba, C.N.; Stanford, K.; Niu, Y.D.; Wang, Y.; McAllister, T.A. Activity of Bacteriophage and Complex Tannins against Biofilm-Forming Shiga Toxin-Producing Escherichia coli from Canada and South Africa. Antibiotics 2020, 9, 257. [Google Scholar] [CrossRef]
- Kim, J.W.; Siletzky, R.M.; Kathariou, S. Host ranges of Listeria-specific bacteriophages from the Turkey processing plant environment in the United States. Appl. Environ. Microbiol. 2008, 74, 6623–6630. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.L.; Hudson, L.K.; Song, Y.; Denes, T.G. Complete Genome Sequences of Two Listeria Phages of the Genus Pecentumvirus. Dennehy, J.J., Ed. Microbiol. Resour. Announc. 2019, 8, 1229–1248. [Google Scholar] [CrossRef] [PubMed]
- Denes, T.; Vongkamjan, K.; Ackermann, H.-W.; Switt, A.I.M.; Wiedmann, M.; den Bakker, H.C. Comparative Genomic and Morphological Analyses of Listeria Phages Isolated from Farm Environments. Appl. Environ. Microbiol. 2014, 80, 4616. [Google Scholar] [CrossRef] [PubMed]
- Blanco Fernandez, M.D.; Klumpp, J.; Barrios, M.E.; Mbayed, V.A. Gene gain and loss and recombination shape evolution of Listeria bacteriophages of the genus Pecentumvirus. Genomics 2021, 113, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, J.; Dorscht, J.; Lurz, R.; Bielmann, R.; Wieland, M.; Zimmer, M.; Calendar, R.; Loessner, M.J. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: A model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 2008, 190, 5753–5765. [Google Scholar] [CrossRef]
- Stone, E.; Lhomet, A.; Neve, H.; Grant, I.R.; Campbell, K.; McAuliffe, O. Isolation and Characterization of Listeria monocytogenes Phage vB_LmoH_P61, a Phage With Biocontrol Potential on Different Food Matrices. Front. Sustain. Food Syst. 2020. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez, M.; Szewczyk, A.; Szamosi, J.; Leung, V.; Filipe, C.; Hosseinidoust, Z. Stress Exposure of Evolved Bacteriophages under Laboratory versus Food Processing Conditions Highlights Challenges in Translatability. Viruses 2023, 15, 113. https://doi.org/10.3390/v15010113
Gomez M, Szewczyk A, Szamosi J, Leung V, Filipe C, Hosseinidoust Z. Stress Exposure of Evolved Bacteriophages under Laboratory versus Food Processing Conditions Highlights Challenges in Translatability. Viruses. 2023; 15(1):113. https://doi.org/10.3390/v15010113
Chicago/Turabian StyleGomez, Mellissa, Alexandra Szewczyk, Jake Szamosi, Vincent Leung, Carlos Filipe, and Zeinab Hosseinidoust. 2023. "Stress Exposure of Evolved Bacteriophages under Laboratory versus Food Processing Conditions Highlights Challenges in Translatability" Viruses 15, no. 1: 113. https://doi.org/10.3390/v15010113
APA StyleGomez, M., Szewczyk, A., Szamosi, J., Leung, V., Filipe, C., & Hosseinidoust, Z. (2023). Stress Exposure of Evolved Bacteriophages under Laboratory versus Food Processing Conditions Highlights Challenges in Translatability. Viruses, 15(1), 113. https://doi.org/10.3390/v15010113