Fast and Sensitive Detection of Soil-Borne Cereal Mosaic Virus in Leaf Crude Extract of Durum Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Samples
2.2. Sample Preparation
2.2.1. RNA Extraction and cDNA Synthesis
2.2.2. Crude Extract Preparation
2.3. SBCMV Detection
2.3.1. LAMP Primer Design
2.3.2. LAMP and Reverse Transcription LAMP (RT-LAMP)
2.3.3. Real-Time PCR Primer Design
2.3.4. Real-Time PCR Reaction
2.3.5. Sensitivity Comparison between LAMP and Real-Time PCR Assays
3. Results
3.1. Development of LAMP Protocol
3.1.1. Choice of Primers and Optimization of Reaction Temperature
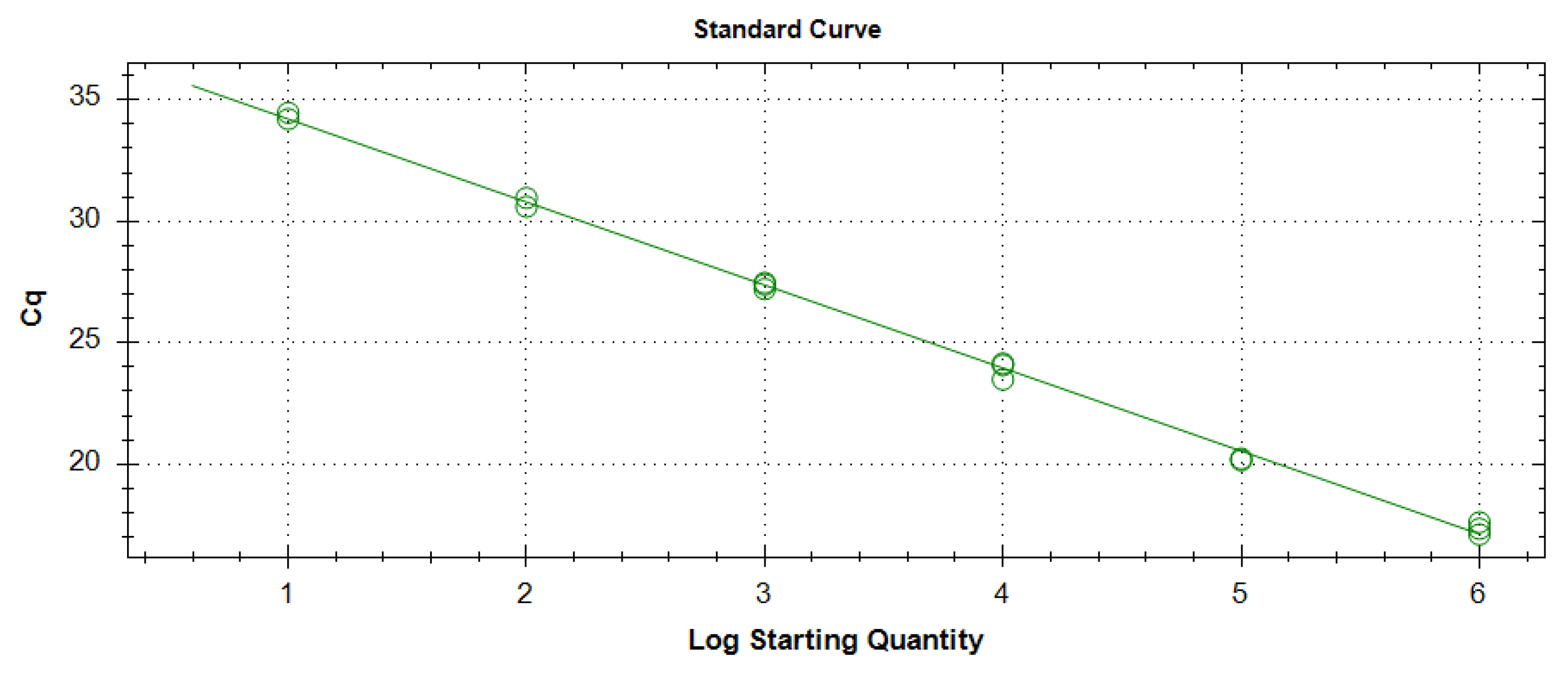
3.1.2. Testing the LAMP Protocol on Different Dilutions of Plant-Derived cDNA and Comparison with Real-Time PCR
3.1.3. Protocol Specificity
3.2. RT-LAMP Protocol Optimisation on Crude Leaf Extracts
3.2.1. Test of Three Different Buffers
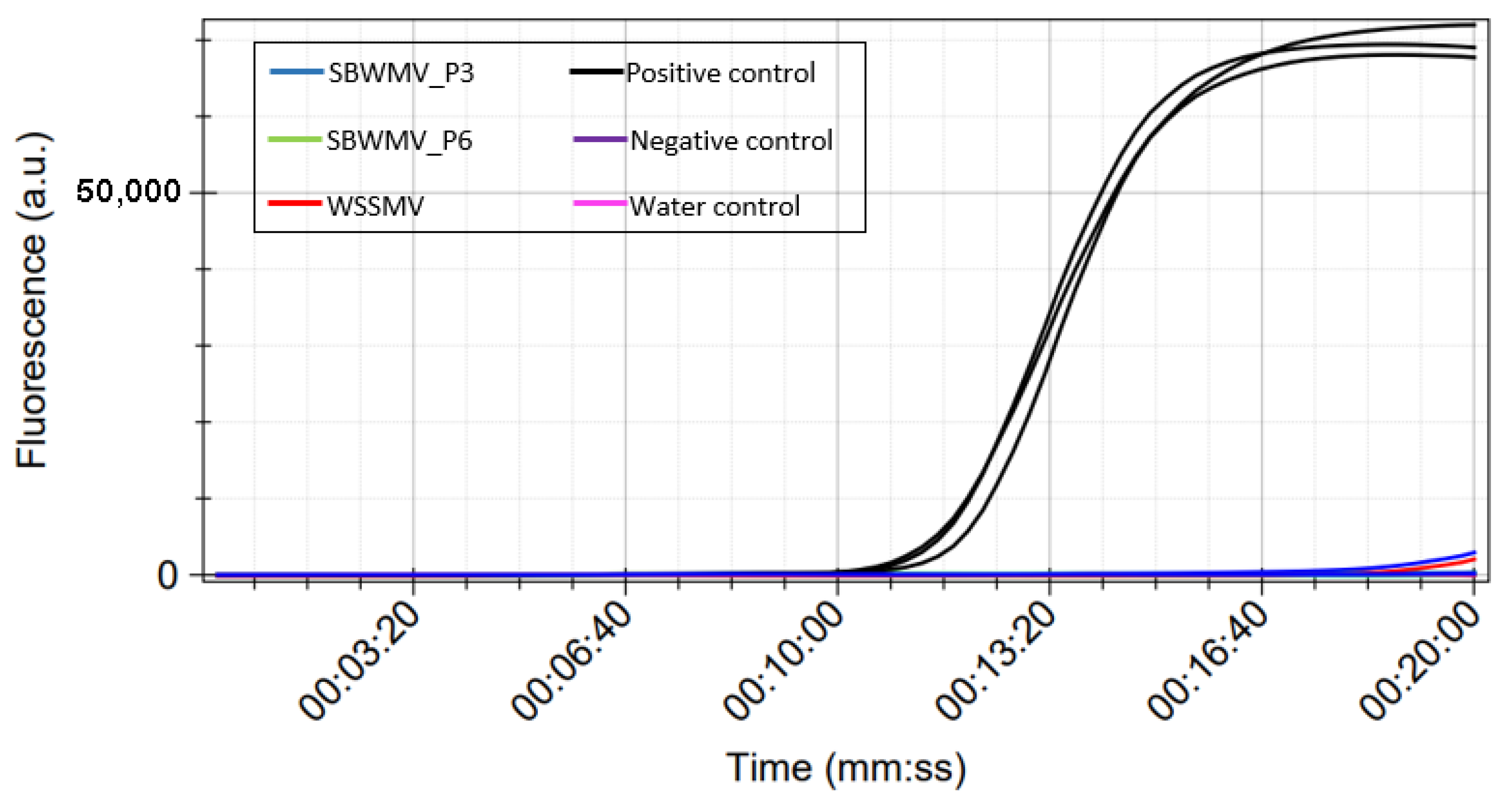
3.2.2. Detection of SBCMV in Crude Extracts
3.2.3. Optimization of the RT-LAMP Reaction by Avian Myeloblastosis Virus Reverse Transcriptase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EUROSTAT. 2022. Available online: https://ec.europa.eu/eurostat/databrowser/view/EF_LAC_CEREALS__custom_3306203/default/table?lang=en (accessed on 6 September 2022).
- Pasqualone, A.; Alba, V.; Mangini, G.; Blanco, A.; Montemurro, C. Durum Wheat Cultivar Traceability in PDO Altamura Bread by Analysis of DNA Microsatellites. Eur. Food Res. Technol. 2010, 230, 723–729. [Google Scholar] [CrossRef]
- Kühne, T. Soil-Borne Viruses Affecting Cereals—Known for Long but Still a Threat. Virus Res. 2009, 141, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Trębicki, P.; Nancarrow, N.; Cole, E.; Bosque-Pérez, N.A.; Constable, F.E.; Freeman, A.J.; Rodoni, B.; Yen, A.L.; Luck, J.E.; Fitzgerald, G.J. Virus Disease in Wheat Predicted to Increase with a Changing Climate. Glob. Chang. Biol. 2015, 21, 3511–3519. [Google Scholar] [CrossRef] [PubMed]
- Mishchenko, L.; Dunich, A.; Mishchenko, I.; Berlizov, V.; Petrenkova, V.; Molchanets, O. Influence of climate changes on wheat viruses variability in Ukraine. Agric. For. 2017, 63, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [Green Version]
- Rao, A.S.; Brakke, M.K. Relation of Soil-Borne Wheat Mosaic Virus and Its Fungal Vector, Polymyxa graminis. Phytopathology 1969, 59, 581–587. [Google Scholar]
- Huth, W. Die bodenbürtigen Viren von Weizen und Roggen in Europa—Ein zunehmendes aber durch ackerbauliche Maßnahmen und Anbau resistenter Sorten lösbares Problem. Gesunde Pflanz. 2002, 54, 51–57. [Google Scholar] [CrossRef]
- Lapierre, H.D.; Hariri, D. Cereal viruses: Wheat and barley. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., van Regenmortel, M.H.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 490–497. [Google Scholar]
- Budge, G.; Ratti, C.; Rubies-Autonell, C.; Lockley, D.; Bonnefoy, M.; Vallega, V.; Pietravalle, S.; Henry, C. Response of UK Winter Wheat Cultivars to Soil-Borne Cereal Mosaic and Wheat Spindle Streak Mosaic Viruses across Europe. Eur. J. Plant Pathol. 2008, 120, 259–272. [Google Scholar] [CrossRef]
- Ziegler, A.; Fomitcheva, V.; Zakri, A.M.; Kastirr, U. Occurrence of Polymyxa graminis Ribotypes in Germany and Their Association with Different Host Plants and Viruses. Cereal Res. Commun. 2016, 44, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Vallega, V. Reactions of Italian Triticum durum Cultivars to Soilborne Wheat Mosaic. Plant Dis. 1985, 69, 64. [Google Scholar] [CrossRef] [Green Version]
- Campbell, L.G.; Heyne, E.G.; Gronau, D.M.; Niblett, C. Effect of soilborne wheat mosaic on wheat yield. Plant Dis. Rep. 1975, 59, 472–476. [Google Scholar]
- Dalbosco, M.; Schons, J.; Prestes, A.M.; Cecchetti, D. Efeito do vírus do mosaico do trigo sobre o rendimento de trigo e triticale. Fitopatol. Bras. 2002, 27, 53–57. [Google Scholar] [CrossRef]
- Kanyuka, K.; Ward, E.; Adams, M.J. Polymyxa graminis and the Cereal Viruses It Transmits: A Research Challenge. Mol. Plant Pathol. 2003, 4, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Slykhuis, J.T.; Barr, D.J.S. Confirmation of Polymyxa graminis as a vector of Wheat spindle streak mosaic virus. Phytopathology 1978, 68, 639–643. [Google Scholar] [CrossRef] [Green Version]
- Koenig, R. Furovirus. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Vallega, V.; Ratti, C.; Rubies-Autonell, C. Reaction of Durum Wheat Cultivars to Mixed SBWMV and WSSMV Infection in Central Italy. Phytopathol. Mediterr. 2003, 42, 149–154. [Google Scholar] [CrossRef]
- Koenig, R.; Huth, W. Natural Infection of Wheat by the Type Strain of Soil-Borne Wheat Mosaic Virus in a Field in Southern Germany. Eur. J. Plant Pathol. 2003, 109, 191–193. [Google Scholar] [CrossRef]
- Fomitcheva, V.; Kastirr, U.; Habekuss, A.; Kűhne, T. Diagnostic multiplex RT-PCR analysis for the detection of soil-borne viruses and their natural vector Polymyxa graminis. In Proceedings of the 7th Symposium of the International Working Group on Plant Viruses with Fungal Vectors, 1–4 September 2008; Rush, C.M., Ed.; Julius Kühn-Institute Federal Research Centre for Cultivated Plants: Quedlinburg, Germany, 2008; pp. 43–48. [Google Scholar]
- Jeżewska, M.; Trzmiel, K. Studies on Cereal Soil-Borne Viruses in Poland. J. Plant Prot. Res. 2010, 50, 527–534. [Google Scholar] [CrossRef]
- Ratti, C.; Budge, G.; Ward, L.; Clover, G.; Rubies-Autonell, C.; Henry, C. Detection and Relative Quantitation of Soil-Borne Cereal Mosaic Virus (SBCMV) and Polymyxa graminis in Winter Wheat Using Real-Time PCR (TaqMan®). J. Virol. Methods 2004, 122, 95–103. [Google Scholar] [CrossRef]
- Kanyuka, K.; Lovell, D.J.; Mitrofanova, O.P.; Hammond-Kosack, K.; Adams, M.J. A Controlled Environment Test for Resistance to Soil-Borne Cereal Mosaic Virus (SBCMV) and Its Use to Determine the Mode of Inheritance of Resistance in Wheat Cv. Cadenza and for Screening Triticum Monococcum Genotypes for Sources of SBCMV Resistance. Plant Pathol. 2004, 53, 154–160. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [Green Version]
- Panno, S.; Matić, S.; Tiberini, A.; Caruso, A.G.; Bella, P.; Torta, L.; Stassi, R.; Davino, A.S. Loop Mediated Isothermal Amplification: Principles and Applications in Plant Virology. Plants 2020, 9, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, S.F.; Matic, S.; Spadaro, D. Impiego di nuove tecniche molecolari per la diagnosi delle malattie. Prot. Colt. 2018, 2, 9–14. [Google Scholar]
- Waliullah, S.; Ling, K.-S.; Cieniewicz, E.J.; Oliver, J.E.; Ji, P.; Ali, M.E. Development of Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Cucurbit Leaf Crumple Virus. Int. J. Mol. Sci. 2020, 21, 1756. [Google Scholar] [CrossRef] [PubMed]
- Salazar, A.; Ochoa-Corona, F.M.; Olson, J.D.; Babu, B.; Paret, M. Probing Loop-Mediated Isothermal Amplification (LAMP) Targeting Two Gene-Fragments of Rose Rosette Virus. PloS ONE 2021, 16, e0256510. [Google Scholar] [CrossRef]
- Li, X.; Hu, W.; Li, Y.; Li, Y.; Chen, S.; Wang, J. Development of an RT-LAMP Assay for the Detection of Maize Yellow Mosaic Virus in Maize. J. Virol. Methods 2022, 300, 114384. [Google Scholar] [CrossRef]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.-J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a Loop-Mediated Isothermal Amplification Reaction for Diagnostic Applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Özay, B.; McCalla, S.E. A Review of Reaction Enhancement Strategies for Isothermal Nucleic Acid Amplification Reactions. Sens. Actuators Rep. 2021, 3, 100033. [Google Scholar] [CrossRef]
- Romero Romero, J.L.; Carver, G.D.; Arce Johnson, P.; Perry, K.L.; Thompson, J.R. A Rapid, Sensitive and Inexpensive Method for Detection of Grapevine Red Blotch Virus without Tissue Extraction Using Loop-Mediated Isothermal Amplification. Arch. Virol. 2019, 164, 1453–1457. [Google Scholar] [CrossRef]
- Bertacca, S.; Caruso, A.G.; Trippa, D.; Marchese, A.; Giovino, A.; Matic, S.; Noris, E.; Ambrosio, M.I.F.S.; Alfaro, A.; Panno, S.; et al. Development of a Real-Time Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Olea Europaea Geminivirus. Plants 2022, 11, 660. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, T.; Drago, S.; Valentini, F.; Elbeaino, T.; Stampone, G.; Digiaro, M.; D’Onghia, A.M. On-Site Detection of Xylella fastidiosa in Host Plants and in ”spy Insects” Using the Real-Time Loop-Mediated Isothermal Amplification Method. Phytopathol. Mediterr. 2015, 54, 488–496. [Google Scholar]
- Fukuta, S.; Tamura, M.; Maejima, H.; Takahashi, R.; Kuwayama, S.; Tsuji, T.; Yoshida, T.; Itoh, K.; Hashizume, H.; Nakajima, Y.; et al. Differential Detection of Wheat Yellow Mosaic Virus, Japanese Soil-Borne Wheat Mosaic Virus and Chinese Wheat Mosaic Virus by Reverse Transcription Loop-Mediated Isothermal Amplification Reaction. J. Virol. Methods 2013, 189, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kadlubar, F.F.; Chen, J.Z. DNA Supercoiling Suppresses Real-Time PCR: A New Approach to the Quantification of Mitochondrial DNA Damage and Repair. Nucleic Acids Res. 2007, 35, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Shirima, R.R.; Maeda, D.G.; Kanju, E.; Ceasar, G.; Tibazarwa, F.I.; Legg, J.P. Absolute Quantification of Cassava Brown Streak Virus MRNA by Real-Time QPCR. J. Virol. Methods 2017, 245, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gutierrez, S.V.; Goodwin, S.B. Loop-Mediated Isothermal Amplification for Detection of Plant Pathogens in Wheat (Triticum aestivum). Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Y.; Liu, X.-J.; Li, D.-W.; Yu, J.-L.; Han, C.-G. Rapid Detection of Wheat Yellow Mosaic Virus by Reverse Transcription Loop-Mediated Isothermal Amplification. Virol. J. 2011, 8, 550. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kim, J.-H.; Choi, J.-Y.; Jang, W.-C. Loop-Mediated Isothermal Amplification Assay to Rapidly Detect Wheat Streak Mosaic Virus in Quarantined Plants. Plant Pathol. J. 2015, 31, 438–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trzmiel, K.; Hasiów-Jaroszewska, B. Development of Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Genetically Different Wheat Dwarf Virus Isolates. Mol. Biol. Rep. 2020, 47, 8325–8329. [Google Scholar] [CrossRef]


| Primer Set | Name | Sequence (5′–3′) | Genome Position | Concentration Used in LAMP Reaction |
|---|---|---|---|---|
| 1 | F3_1 | CAATCGAAAGTGGTTGTGC | 298–316 | 0.25 μM |
| B3_1 | AGCATTACACCTAAATGGGTA | 459–479 | 0.25 μM | |
| FIP_1 | ATGCCATCAAATTCAATTCCTTGTT-AGTTACCTCAAATGGCTGT | 358–382 (F1c_1) + 317–335 (F2_1) | 2.5 μM | |
| BIP_1 | CTATTCATCCTTTCATTAGGCTGGG-GCTAGCTCGTGTTGCTTG | 383–417 (B1c_1) + 436–453 (B2_1) | 2.5 μM | |
| 2 | F3_2 | GTTACCTCAAATGGCTGTC | 318–336 | 0.25 μM |
| B3_2 | AGAAATTTCGCTCACCTAAC | 495–514 | 0.25 μM | |
| FIP_2 | CCAGCCTAATGAAAGGATGAATAGA-AAGAACGGTTATACGGGTT | 382–406 (F1c_2) + 337–355 (F2_2) | 2.5 μM | |
| BIP_2 | TTGATAAGTCAGATTGAGGGATGGC-ACCGTTGAGCATTACACC | 412–436 (B1c_2) + 469–486 (B2_2) | 2.5 μM | |
| LOOPB_2 | AAGCAACACGAGCTAGCATACTTAC | 437–461 | 1.25 μM |
| Primer Name | Sequence (5′–3′) | Genome Position | Fragment Size (nt) |
|---|---|---|---|
| qPCR_577fw | GGWGGTGARGCAGTTATGC | 577–595 | 137 |
| qPCR_714rv | ACCYTGYTCCTCACCCTCAA | 695–714 | |
| AJ132577_157fw_CP | GGTAGTCAGCTGTTAGCGTGT | 157–177 | 773 |
| AJ132577_929rv_CP | TCGGCCAAAACCAGCCTATT | 910–929 |
| Sample | cDNA Dilution | LAMP |
|---|---|---|
| Positive 1 | 10−3 | +/+/+ |
| 10−4 | +/+/+ | |
| 10−5 | +/+/+ | |
| 10−6 | +/+/+ | |
| 10−7 | +/−/− | |
| Positive 2 | 10−3 | +/+/+ |
| 10−4 | +/+/+ | |
| 10−5 | +/+/+ | |
| 10−6 | +/+/+ | |
| 10−7 | +/+/+ | |
| Positive 3 | 10−3 | +/+/+ |
| 10−4 | +/+/+ | |
| 10−5 | +/+/+ | |
| 10−6 | +/+/+ | |
| 10−7 | +/+/− | |
| Negative | 10−3 | −/−/− |
| 10−4 | −/−/− | |
| 10−5 | −/−/− | |
| 10−6 | −/−/− | |
| 10−7 | −/−/− |
| Sample | LAMP | Real-Time PCR | ||
|---|---|---|---|---|
| (cDNA) | Rt (min) ± SD | Tm (°C) ± SD | Cq ± SD | Tm (°C) ± SD |
| 1_A | nd | nd | nd | nd |
| 1_B | nd | nd | nd | nd |
| 1_C | nd | nd | nd | nd |
| 5_A | nd | nd | nd | nd |
| 5_B | 8.6 ± 0.7 | 84.5 ± 0.0 | 30.06 ± 0.26 | 80.5 ± 0.0 |
| 5_C | 6.13 ± 0.06 | 84.5 ± 0.0 | 22.8 ± 0.3 | 80.17 ± 0.29 |
| 166_A | nd | nd | nd | nd |
| 166_B | nd | nd | nd | nd |
| 166_C | nd | nd | nd | nd |
| 171_A | nd | nd | nd | nd |
| 171_B | nd | nd | nd | nd |
| 171_C | 8.25 ± 0.10 | 84.5 ± 0.0 | 29.55 ± 0.27 | 80.5 ± 0.0 |
| 209_A | 6.81 ± 0.13 | 84.5 ± 0.0 | 25.46 ± 0.26 | 80.5 ± 0.0 |
| 209_B | nd | nd | nd | nd |
| 209_C | nd | nd | nd | nd |
| 210_A | nd | nd | nd | nd |
| 210_B | nd | nd | nd | nd |
| 210_C | nd | nd | nd | nd |
| 213_A | nd | nd | nd | nd |
| 213_B | nd | nd | nd | nd |
| 213_C | nd | nd | nd | nd |
| 214_A | nd | nd | nd | nd |
| 214_B | nd | nd | nd | nd |
| 214_C | nd | nd | nd | nd |
| Positive control | 9.40 ± 0.22 | 84.5 ± 0.0 | 18.238 ± 0.026 | 80.17 ± 0.29 |
| Negative control | nd | nd | nd | nd |
| Sample | Diluition | LAMP Tp (min) ± SD | qPCR Cq ± SD | qPCR Quantification EC ± SD |
|---|---|---|---|---|
| Positive 1 | 10−3 | 5.327 ± 0.029 | 22.70 ± 0.15 | 24,700 ± 2500 |
| 10−4 | 6.28 ± 0.18 | 26.02 ± 0.13 | 2720 ± 220 | |
| 10−5 | 7.77 ± 0.19 | (29.71 ± 0.21) ** | 240 ± 30 | |
| 10−6 | 9.2 ± 0.9 | (32.13 ± 0.06) ** | 47.2 ± 1.8 | |
| 10−7 | nd * | nd | nd | |
| Positive 2 | 10−3 | 4.90 ± 0.15 | 21.357 ± 0. 017 | 60,000 ± 700 |
| 10−4 | 5.60 ± 0.06 | 24.277 ± 0.025 | 8650 ± 140 | |
| 10−5 | 7.08 ± 0.19 | 27.5 ± 0.4 | 1120 ± 240 | |
| 10−6 | 8.6 ± 1.3 | 29.97 ± 0.11 | 199 ± 14 | |
| 10−7 | 9.6 ± 1.4 | nd | nd | |
| Positive 3 | 10−3 | 4.99 ± 0.16 | 21.59 ± 0.09 | 51,000 ± 3000 |
| 10−4 | 5.84 ± 0.05 | 24.9 ± 0.4 | 5900 ± 1400 | |
| 10−5 | 6.87 ± 0.17 | 27.90 ± 0.16 | 780 ± 90 | |
| 10−6 | 9.6 ± 0.6 | 30.6 ± 0.4 | 130 ± 30 | |
| 10−7 | (9.3 ± 0.4) ** | nd | nd | |
| Negative | 10−3 | nd | nd | nd |
| 10−4 | nb | nb | nd | |
| 10−5 | nb | nb | nd | |
| 10−6 | nb | nb | nd | |
| 10−7 | nb | nb | nd |
| Sample | Dilution | TET Rt (min) ± SD | Tris HCl Rt (min) ± SD |
|---|---|---|---|
| 1:103 | 11.4 ± 0.7 | 12.4 ± 0.5 | |
| Positive control | 1:104 | 13.00 ± 0.29 | 13.6 ± 1.5 |
| 1:105 | 13.4 ± 0.8 | 15.2 ± 0.8 | |
| 1:103 | nd | nd | |
| Negative control | 1:104 | nd | nd |
| 1:105 | nd | nd | |
| Water control | - | nd | nd |
| Sample (Crude Extract) | RT-LAMP Rt (min) ± SD | RT-LAMP with AMV-RT Rt (min) ± SD |
|---|---|---|
| 1A | nd | nt |
| 1B | nd | nt |
| 1C | nd | nt |
| 5A | nd | nd |
| 5B | nd | 6.99 ± 0.10 |
| 5C | nd | 6.49 ± 0.08 |
| 166A | nd | nt |
| 166B | nd | nt |
| 166C | nd | nt |
| 171A | nd | nd |
| 171B | nd | nd |
| 171C | nd | 7.43 ± 0.23 |
| 209A | 10.5 ± 0.8 | 6.62 ± 0.03 |
| 209B | nd | nd |
| 209C | nd | nd |
| 210A | nd | nd |
| 210B | nd | nd |
| 210C | nd | nd |
| 213A | nd | nt |
| 213B | nd | nt |
| 213C | nd | nt |
| 214A | nd | nt |
| 214B | nd | nt |
| 214C | nd | nt |
| Positive | (11.6 ± 1.2) * | (6.1 ± 0.6) ** |
| Negative | nd | nd |
| Water | nd | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marra, M.; D’Errico, C.; Montemurro, C.; Ratti, C.; Baldoni, E.; Matic, S.; Accotto, G.P. Fast and Sensitive Detection of Soil-Borne Cereal Mosaic Virus in Leaf Crude Extract of Durum Wheat. Viruses 2023, 15, 140. https://doi.org/10.3390/v15010140
Marra M, D’Errico C, Montemurro C, Ratti C, Baldoni E, Matic S, Accotto GP. Fast and Sensitive Detection of Soil-Borne Cereal Mosaic Virus in Leaf Crude Extract of Durum Wheat. Viruses. 2023; 15(1):140. https://doi.org/10.3390/v15010140
Chicago/Turabian StyleMarra, Monica, Chiara D’Errico, Cinzia Montemurro, Claudio Ratti, Elena Baldoni, Slavica Matic, and Gian Paolo Accotto. 2023. "Fast and Sensitive Detection of Soil-Borne Cereal Mosaic Virus in Leaf Crude Extract of Durum Wheat" Viruses 15, no. 1: 140. https://doi.org/10.3390/v15010140











