Understanding the Biology and Immune Pathogenesis of Chikungunya Virus Infection for Diagnostic and Vaccine Development
Abstract
:1. Introduction
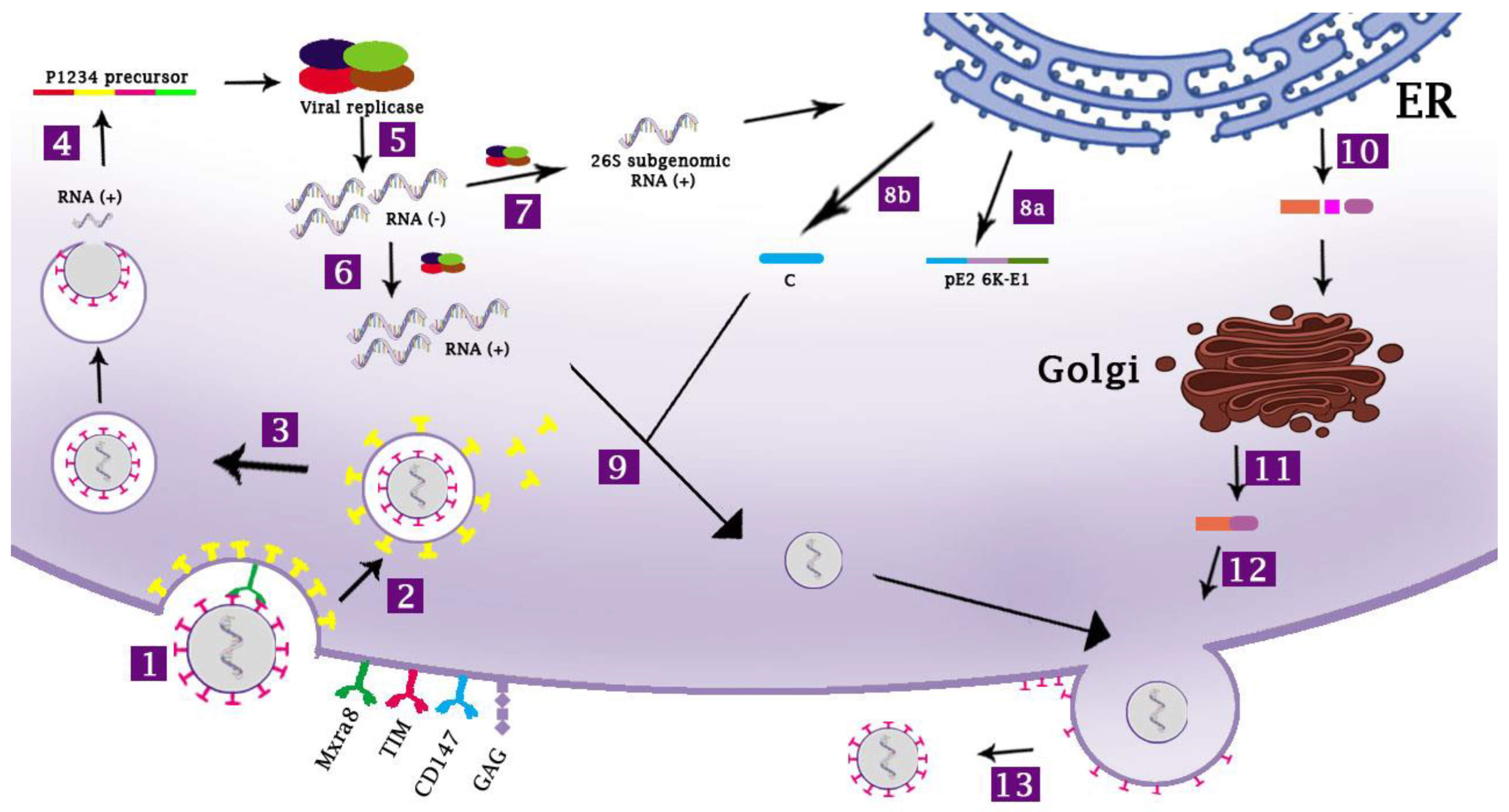
2. The Life Cycle of CHIKV
2.1. The Host Cell Receptors for CHIKV
2.2. The Entry Process of CHIKV
2.3. CHIKV Replication within the Target Cells
3. The Viral and Host Factors Involved in the Pathogenesis of CHIKV Infection
4. The Antiviral IFN Responses against CHIKV Infection
4.1. Stimulation of IFN Responses
4.2. Antagonism of IFN Responses by CHIKV
5. The Host Adaptive Immune Responses against CHIKV Infection
5.1. The T Cell-Mediated Immune Responses against CHIKV
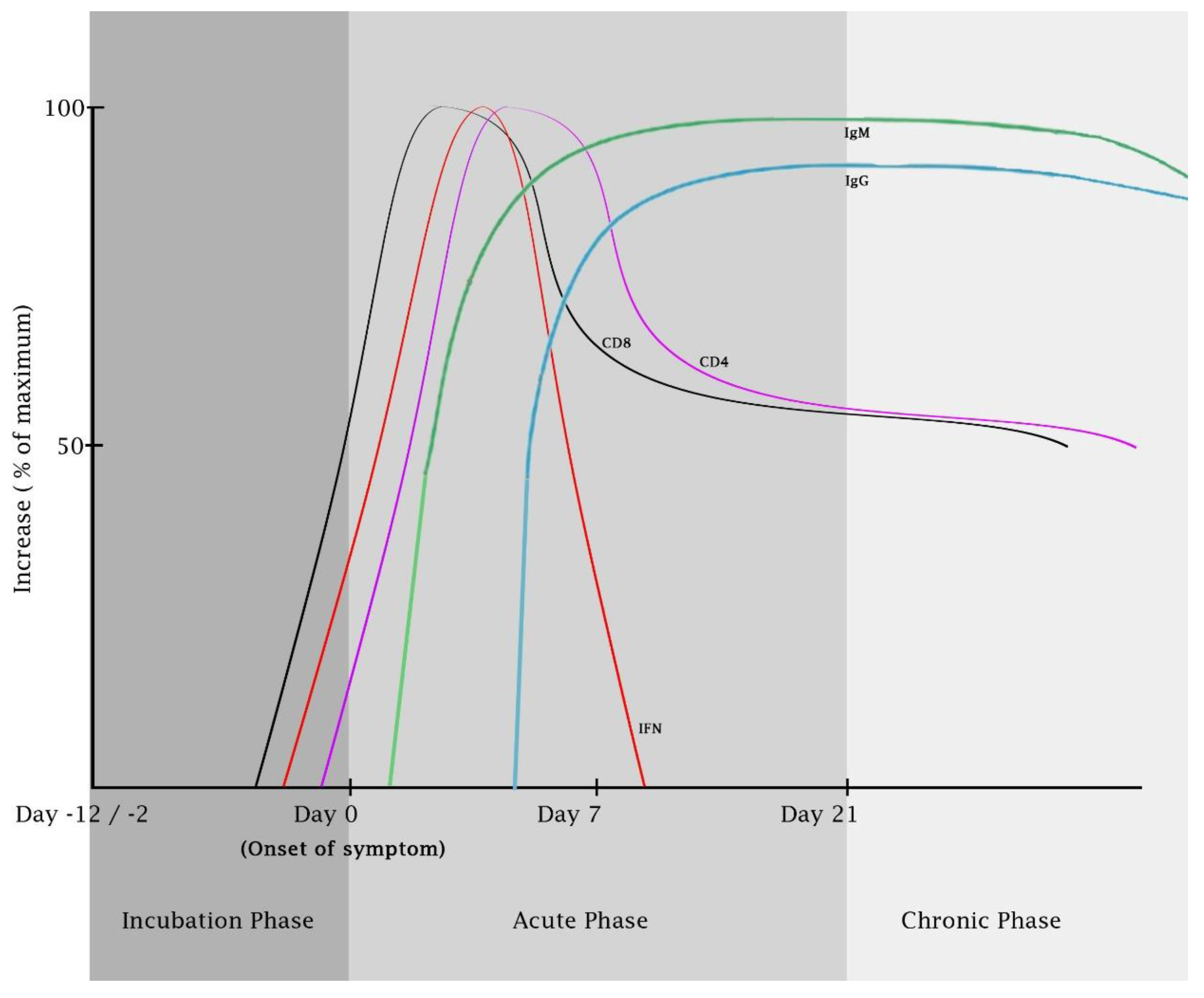
5.2. The Humoral Immune Responses against CHIKV
6. The Clinical Manifestations of CHIKV Infection
6.1. Acute Phase of Infection
6.2. Chronic or Persistent Phase of Infection
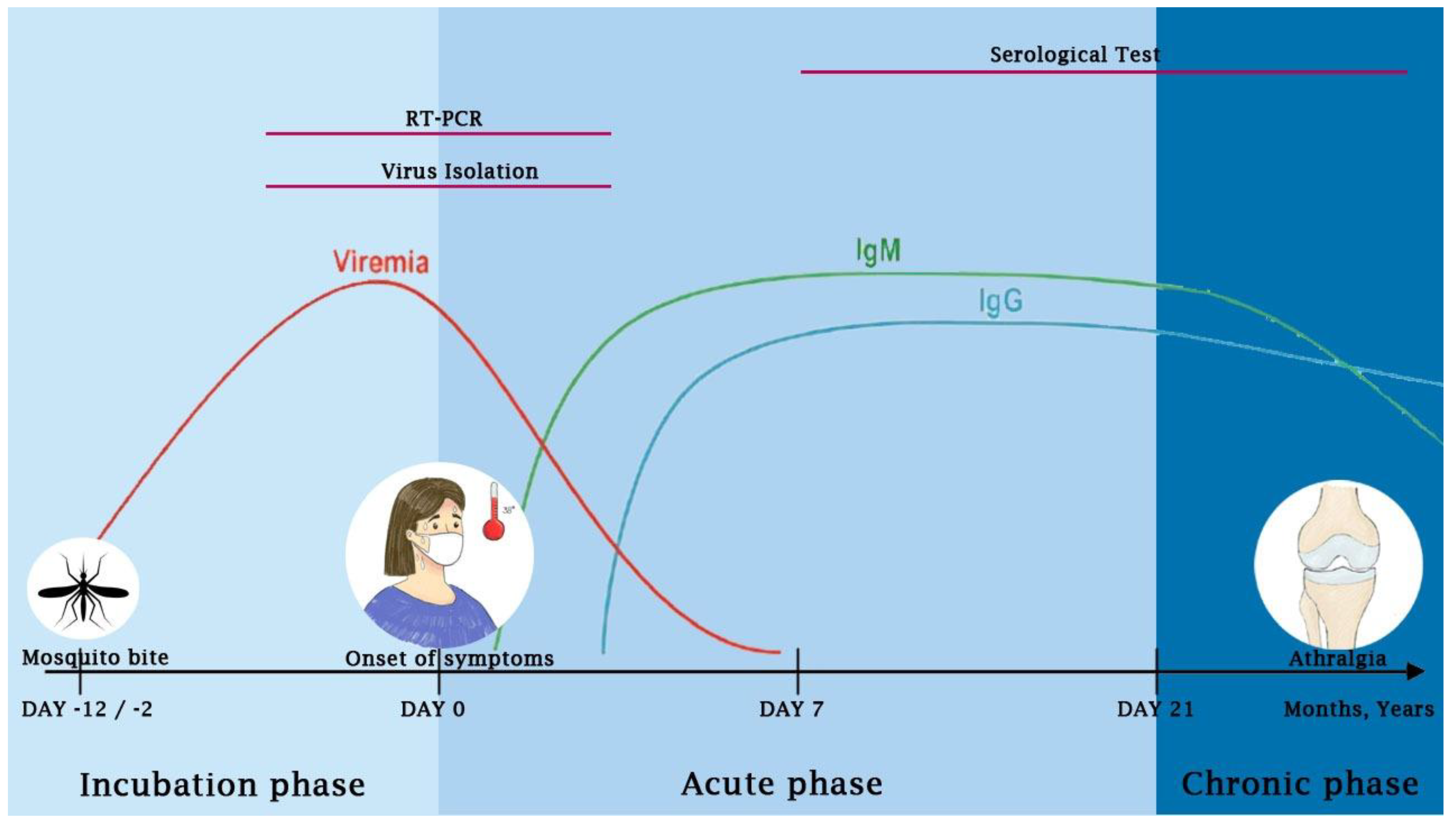
7. Diagnosis of CHIKV Infection
7.1. Virus Isolation
7.2. Nucleic Acid Detection
7.3. Serology Tests
7.3.1. IgM and IgG Antibodies-Based Serological Tests
7.3.2. Antigen-Based Serological Tests
8. The Current Development of Vaccine Candidates against CHIKV
| Vaccine | Platform | Background CHIKV Strain | Number of Doses | The Last Stage of Development | Developer | References |
|---|---|---|---|---|---|---|
| VLA1553 | Live-attenuated | LR2006-OPY1 (ECSA-IOL) | Single dose | Phase III (completed) | Valneva, Austria GmbH | NCT04786444, NCT04838444, NCT04546724, NCT04650399 |
| VRC-CHKVLP059-00-VP | Virus-like particle | 37997 strain of the WA genotype | Two doses | Phase II | The US National Institute of Allergy and Infectious Disease (US NIAID) | NCT02562482, NCT01489358, and [192,193] |
| PXVX0317 | Virus-like particle (adjuvanted) | 37997 strain of the WA genotype | Single dose | Phase III | Emergent BioSolutions (Gaithersburg, MD, USA) | NCT03483961, NCT05072080, NCT05349617, and [194] |
| MV-CHIK-202 | Recombinant measles virus-vectored vaccine | pTM-MVSchw-CE3E26KE1 | Single or two doses | Phase II | Themis Bioscience GmbH | NCT02861586, NCT03101111, and [195,196] |
| BBV87 | Whole virus-inactivated | ECSA | Two doses | Phase II and III | International Vaccine Institute | NCT04566484 |
8.1. Live-Attenuated Viral Vaccines (LAV)
8.2. Nucleic Acid-Based (RNA) Vaccines
8.3. Virus-like Particle (VLPs)
8.4. Recombinant Virus-Vectored Vaccines
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Burt, F.J.; Chen, W.; Miner, J.J.; Lenschow, D.J.; Merits, A.; Schnettler, E.; Kohl, A.; Rudd, P.A.; Taylor, A.; Herrero, L.J.; et al. Chikungunya virus: An update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017, 17, e107–e117. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Tauran, P.; Kosasih, H.; Pelupessy, N.M.; Sennang, N.; Mubin, R.H.; Sudarmono, P.; Tjitra, E.; Murniati, D.; Alam, A.; et al. Chikungunya in Indonesia: Epidemiology and diagnostic challenges. PLoS Negl. Trop. Dis. 2020, 14, e0008355. [Google Scholar] [CrossRef]
- Zaid, A.; Burt, F.J.; Liu, X.; Poo, Y.S.; Zandi, K.; Suhrbier, A.; Weaver, S.C.; Texeira, M.M.; Mahalingam, S. Arthritogenic alphaviruses: Epidemiological and clinical perspective on emerging arboviruses. Lancet Infect. Dis. 2021, 21, e123–e133. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.W. The Newala epidemic. III. The virus: Isolation, pathogenic properties and relationship to the epidemic. J. Hyg. 1956, 54, 177–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.H.; Morita, K.; Parquet, M.D.C.; Hasebe, F.; Mathenge, E.G.M.; Igarashi, A. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J. Gen. Virol. 2002, 83, 3075–3084. [Google Scholar] [CrossRef]
- Sahadeo, N.S.D.; Allicock, O.M.; De Salazar, P.M.; Auguste, A.J.; Widen, S.; Olowokure, B.; Gutierrez, C.; Valadere, A.M.; Polson-Edwards, K.; Weaver, S.C.; et al. Understanding the evolution and spread of chikungunya virus in the Americas using complete genome sequences. Virus Evol. 2017, 3, vex010. [Google Scholar] [CrossRef] [Green Version]
- Volk, S.M.; Chen, R.; Tsetsarkin, K.A.; Adams, A.P.; Garcia, T.I.; Sall, A.A.; Nasar, F.; Schuh, A.J.; Holmes, E.C.; Higgs, S.; et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J. Virol. 2010, 84, 6497–6504. [Google Scholar] [CrossRef] [Green Version]
- Zeller, H.; Van Bortel, W.; Sudre, B. Chikungunya: Its history in Africa and Asia and its spread to new regions in 2013–2014. J. Infect. Dis. 2016, 214, S436–S440. [Google Scholar] [CrossRef]
- Petersen, L.R.; Powers, A.M. Chikungunya: Epidemiology. F1000Res 2016, 5, F1000 Faculty Rev-1082. [Google Scholar] [CrossRef] [Green Version]
- Pathak, H.; Mohan, M.C.; Ravindran, V. Chikungunya arthritis. Clin. Med. 2019, 19, 381–385. [Google Scholar] [CrossRef]
- Weber, C.; Berberich, E.; von Rhein, C.; Henss, L.; Hildt, E.; Schnierle, B.S. Identification of functional determinants in the chikungunya virus E2 protein. PLoS Negl. Trop. Dis. 2017, 11, e0005318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Zhao, Z.; Chai, Y.; Jin, X.; Li, C.; Yuan, F.; Liu, S.; Gao, Z.; Wang, H.; Song, J.; et al. Molecular basis of arthritogenic alphavirus receptor MXRA8 binding to chikungunya virus envelope protein. Cell 2019, 177, 1714–1724.e1712. [Google Scholar] [CrossRef] [PubMed]
- Basore, K.; Kim, A.S.; Nelson, C.A.; Zhang, R.; Smith, B.K.; Uranga, C.; Vang, L.; Cheng, M.; Gross, M.L.; Smith, J.; et al. Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor. Cell 2019, 177, 1725–1737.e1716. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Du, X.; Cheng, W. Structures unveil the invasion mechanism of chikungunya virus. Trends Microbiol. 2019, 27, 656–658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Earnest, J.T.; Kim, A.S.; Winkler, E.S.; Desai, P.; Adams, L.J.; Hu, G.; Bullock, C.; Gold, B.; Cherry, S.; et al. Expression of the Mxra8 receptor promotes alphavirus infection and pathogenesis in mice and drosophila. Cell Rep. 2019, 28, 2647–2658.e2645. [Google Scholar] [CrossRef]
- Zhang, R.; Kim, A.S.; Fox, J.M.; Nair, S.; Basore, K.; Klimstra, W.B.; Rimkunas, R.; Fong, R.H.; Lin, H.; Poddar, S.; et al. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 2018, 557, 570–574. [Google Scholar] [CrossRef]
- Kim, A.S.; Zimmerman, O.; Fox, J.M.; Nelson, C.A.; Basore, K.; Zhang, R.; Durnell, L.; Desai, C.; Bullock, C.; Deem, S.L.; et al. An evolutionary insertion in the Mxra8 receptor-binding site confers resistance to alphavirus infection and pathogenesis. Cell Host Microbe 2020, 27, 428–440.e429. [Google Scholar] [CrossRef]
- Yin, P.; Kielian, M. BHK-21 cell clones differ in chikungunya virus infection and MXRA8 receptor expression. Viruses 2021, 13, 949. [Google Scholar] [CrossRef]
- Powell, L.A.; Miller, A.; Fox, J.M.; Kose, N.; Klose, T.; Kim, A.S.; Bombardi, R.; Tennekoon, R.N.; Dharshan de Silva, A.; Carnahan, R.H.; et al. Human mAbs broadly protect against arthritogenic aAlphaviruses by recognizing conserved elements of the Mxra8 receptor-binding site. Cell Host Microbe 2020, 28, 699–711.e697. [Google Scholar] [CrossRef]
- Kumar, A.; Rathi, E.; Kini, S.G. Exploration of small-molecule entry disruptors for chikungunya virus by targeting matrix remodelling associated protein. Res. Pharm. Sci. 2020, 15, 300–311. [Google Scholar] [CrossRef]
- Verma, J.; Subbarao, N. In silico identification of small molecule protein-protein interaction inhibitors: Targeting hotspot regions at the interface of MXRA8 and CHIKV envelope protein. J. Biomol. Struct. Dyn. 2022. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- De Caluwe, L.; Coppens, S.; Vereecken, K.; Daled, S.; Dhaenens, M.; Van Ostade, X.; Deforce, D.; Arien, K.K.; Bartholomeeusen, K. The CD147 protein complex is involved in entry of chikungunya virus and related alphaviruses in human cells. Front. Microbiol. 2021, 12, 615165. [Google Scholar] [CrossRef] [PubMed]
- McAllister, N.; Liu, Y.; Silva, L.M.; Lentscher, A.J.; Chai, W.; Wu, N.; Griswold, K.A.; Raghunathan, K.; Vang, L.; Alexander, J.; et al. Chikungunya virus strains from each genetic clade bind sulfated glycosaminoglycans as attachment factors. J. Virol. 2020, 94, e01500–e01520. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Tumkosit, U.; Nakamura, S.; Motooka, D.; Kishishita, N.; Priengprom, T.; Sa-Ngasang, A.; Kinoshita, T.; Takeda, N.; Maeda, Y. Genome-wide screening uncovers the significance of N-sulfation of heparan sulfate as a host cell factor for chikungunya virus infection. J. Virol. 2017, 91, e00432-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, L.A.; Khomandiak, S.; Ashbrook, A.W.; Weller, R.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. A single-amino-acid polymorphism in chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J. Virol. 2014, 88, 2385–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, D.; Paul, A.M.; Anderson, J.F.; Huang, F.; Bai, F. Loss of glycosaminoglycan receptor binding after mosquito cell passage reduces chikungunya virus infectivity. PLoS Negl. Trop. Dis. 2015, 9, e0004139. [Google Scholar] [CrossRef] [Green Version]
- Ashbrook, A.W.; Burrack, K.S.; Silva, L.A.; Montgomery, S.A.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. Residue 82 of the chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J. Virol. 2014, 88, 12180–12192. [Google Scholar] [CrossRef] [Green Version]
- Rangel, M.V.; McAllister, N.; Dancel-Manning, K.; Noval, M.G.; Silva, L.A.; Stapleford, K.A. Emerging chikungunya virus variants at the E1-E1 interglycoprotein spike interface impact virus attachment and inflammation. J. Virol. 2022, 96, e0158621. [Google Scholar] [CrossRef]
- Wintachai, P.; Wikan, N.; Kuadkitkan, A.; Jaimipuk, T.; Ubol, S.; Pulmanausahakul, R.; Auewarakul, P.; Kasinrerk, W.; Weng, W.Y.; Panyasrivanit, M.; et al. Identification of prohibitin as a chikungunya virus receptor protein. J. Med. Virol. 2012, 84, 1757–1770. [Google Scholar] [CrossRef]
- Wintachai, P.; Thuaud, F.; Basmadjian, C.; Roytrakul, S.; Ubol, S.; Desaubry, L.; Smith, D.R. Assessment of flavaglines as potential chikungunya virus entry inhibitors. Microbiol. Immunol. 2015, 59, 129–141. [Google Scholar] [CrossRef]
- Kirui, J.; Abidine, Y.; Lenman, A.; Islam, K.; Gwon, Y.D.; Lasswitz, L.; Evander, M.; Bally, M.; Gerold, G. The phosphatidylserine receptor TIM-1 enhances authentic chikungunya virus cell entry. Cells 2021, 10, 1828. [Google Scholar] [CrossRef] [PubMed]
- Hoornweg, T.E.; van Duijl-Richter, M.K.S.; Ayala Nunez, N.V.; Albulescu, I.C.; van Hemert, M.J.; Smit, J.M. Dynamics of chikungunya virus cell entry unraveled by single-virus tracking in living cells. J. Virol. 2016, 90, 4745–4756. [Google Scholar] [CrossRef] [Green Version]
- Bernard, E.; Solignat, M.; Gay, B.; Chazal, N.; Higgs, S.; Devaux, C.; Briant, L. Endocytosis of chikungunya virus into mammalian cells: Role of clathrin and early endosomal compartments. PLoS ONE 2010, 5, e11479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.H.R.; Mohamed Hussain, K.; Chu, J.J.H. Macropinocytosis dependent entry of chikungunya virus into human muscle cells. PLoS Negl. Trop. Dis. 2019, 13, e0007610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumida, M.; Hayashi, H.; Tanaka, A.; Kubo, Y. Cathepsin B protease facilitates chikungunya virus envelope protein-mediated infection via endocytosis or macropinocytosis. Viruses 2020, 12, 722. [Google Scholar] [CrossRef]
- Rausalu, K.; Utt, A.; Quirin, T.; Varghese, F.S.; Zusinaite, E.; Das, P.K.; Ahola, T.; Merits, A. Chikungunya virus infectivity, RNA replication and non-structural polyprotein processing depend on the nsP2 protease’s active site cysteine residue. Sci. Rep. 2016, 6, 37124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freire, M.; Basso, L.G.M.; Mendes, L.F.S.; Mesquita, N.; Mottin, M.; Fernandes, R.S.; Policastro, L.R.; Godoy, A.S.; Santos, I.A.; Ruiz, U.E.A.; et al. Characterization of the RNA-dependent RNA polymerase from chikungunya virus and discovery of a novel ligand as a potential drug candidate. Sci. Rep. 2022, 12, 10601. [Google Scholar] [CrossRef]
- Albulescu, I.C.; Tas, A.; Scholte, F.E.M.; Snijder, E.J.; van Hemert, M.J. An in vitro assay to study chikungunya virus RNA synthesis and the mode of action of inhibitors. J. Gen. Virol. 2014, 95, 2683–2692. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.A.; Dermody, T.S. Chikungunya virus: Epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Investig. 2017, 127, 737–749. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Kesari, P.; Kumar, P.; Tomar, S. Structure-function insights into chikungunya virus capsid protein: Small molecules targeting capsid hydrophobic pocket. Virology 2018, 515, 223–234. [Google Scholar] [CrossRef]
- Thomas, S.; Rai, J.; John, L.; Gunther, S.; Drosten, C.; Putzer, B.M.; Schaefer, S. Functional dissection of the alphavirus capsid protease: Sequence requirements for activity. Virol. J. 2010, 7, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, R.S.; Anastasakis, D.G.; Hafner, M.; Kielian, M. Multiple capsid protein binding sites mediate selective packaging of the alphavirus genomic RNA. Nat. Commun. 2020, 11, 4693. [Google Scholar] [CrossRef]
- Constant, L.E.C.; Rajsfus, B.F.; Carneiro, P.H.; Sisnande, T.; Mohana-Borges, R.; Allonso, D. Overview on chikungunya virus infection: From epidemiology to state-of-the-art experimental models. Front. Microbiol. 2021, 12, 744164. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, C.; Pristatsky, P.; Hoang, V.M.; Casimiro, D.R.; Schwartz, R.M.; Rustandi, R.; Ha, S. Characterization of N-glycosylation profiles from mammalian and insect cell derived chikungunya VLP. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1032, 218–223. [Google Scholar] [CrossRef]
- Yap, M.L.; Klose, T.; Urakami, A.; Hasan, S.S.; Akahata, W.; Rossmann, M.G. Structural studies of chikungunya virus maturation. Proc. Natl. Acad. Sci. USA 2017, 114, 13703–13707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chmielewski, D.; Schmid, M.F.; Simmons, G.; Jin, J.; Chiu, W. Chikungunya virus assembly and budding visualized in situ using cryogenic electron tomography. Nat. Microbiol. 2022, 7, 1270–1279. [Google Scholar] [CrossRef]
- Brown, R.S.; Wan, J.J.; Kielian, M. The alphavirus exit pathway: What we know and what we wish we knew. Viruses 2018, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Sourisseau, M.; Schilte, C.; Casartelli, N.; Trouillet, C.; Guivel-Benhassine, F.; Rudnicka, D.; Sol-Foulon, N.; Le Roux, K.; Prevost, M.C.; Fsihi, H.; et al. Characterization of reemerging chikungunya virus. PLoS. Pathog. 2007, 3, e89. [Google Scholar] [CrossRef]
- Noret, M.; Herrero, L.; Rulli, N.; Rolph, M.; Smith, P.N.; Li, R.W.; Roques, P.; Gras, G.; Mahalingam, S. Interleukin 6, RANKL, and osteoprotegerin expression by chikungunya virus-infected human osteoblasts. J. Infect. Dis. 2012, 206, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Pott, F.; Postmus, D.; Brown, R.J.P.; Wyler, E.; Neumann, E.; Landthaler, M.; Goffinet, C. Single-cell analysis of arthritogenic alphavirus-infected human synovial fibroblasts links low abundance of viral RNA to induction of innate immunity and arthralgia-associated gene expression. Emerg. Microbes Infect. 2021, 10, 2151–2168. [Google Scholar] [CrossRef]
- Hibl, B.M.; Dailey Garnes, N.J.M.; Kneubehl, A.R.; Vogt, M.B.; Spencer Clinton, J.L.; Rico-Hesse, R.R. Mosquito-bite infection of humanized mice with chikungunya virus produces systemic disease with long-term effects. PLoS. Negl. Trop. Dis. 2021, 15, e0009427. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, H.; Li, X.; Zhou, D.; Zhong, M.; Yang, J.; Zhao, B.; Fan, X.; Fan, J.; Shu, J.; et al. A high-dose inoculum size results in persistent viral infection and arthritis in mice infected with chikungunya virus. PLoS. Negl. Trop. Dis. 2022, 16, e0010149. [Google Scholar] [CrossRef] [PubMed]
- Labadie, K.; Larcher, T.; Joubert, C.; Mannioui, A.; Delache, B.; Brochard, P.; Guigand, L.; Dubreil, L.; Lebon, P.; Verrier, B.; et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J. Clin. Investig. 2010, 120, 894–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, J.P.; McCarthy, M.K.; Davenport, B.J.; Morrison, T.E.; Diamond, M.S. Clearance of chikungunya virus infection in lymphoid tissues is promoted by treatment with an agonistic anti-CD137 antibody. J. Virol. 2019, 93, e01231-19. [Google Scholar] [CrossRef] [PubMed]
- Raghavendar, B.S.; Patel, A.K.; Kabra, S.K.; Lodha, R.; Ratageri, V.H.; Ray, P. Virus load and clinical features during the acute phase of chikungunya infection in children. PLoS ONE 2019, 14, e0211036. [Google Scholar] [CrossRef]
- Reddy, V.; Mani, R.S.; Desai, A.; Ravi, V. Correlation of plasma viral loads and presence of chikungunya IgM antibodies with cytokine/chemokine levels during acute chikungunya virus infection. J. Med. Virol. 2014, 86, 1393–1401. [Google Scholar] [CrossRef]
- Imad, H.A.; Phadungsombat, J.; Nakayama, E.E.; Kludkleeb, S.; Matsee, W.; Ponam, T.; Suzuki, K.; Leaungwutiwong, P.; Piyaphanee, W.; Phumratanaprapin, W.; et al. Chikungunya manifestations and viremia in patients who presented to the fever clinic at Bangkok hospital for tropical diseases during the 2019 outbreak in Thailand. Trop. Med. Infect. Dis. 2021, 6, 12. [Google Scholar] [CrossRef]
- Pinzon-Redondo, H.; Paternina-Caicedo, A.; Barrios-Redondo, K.; Zarate-Vergara, A.; Tirado-Perez, I.; Fortich, R.; Alvis-Guzman, N.; Mattar, S. Risk factors for severity of chikungunya in children: A prospective assessment. Pediatr. Infect. Dis. J. 2016, 35, 702–704. [Google Scholar] [CrossRef] [Green Version]
- Waggoner, J.J.; Gresh, L.; Vargas, M.J.; Ballesteros, G.; Tellez, Y.; Soda, K.J.; Sahoo, M.K.; Nunez, A.; Balmaseda, A.; Harris, E.; et al. Viremia and clinical presentation in Nicaraguan patients infected with Zika virus, chikungunya virus, and Dengue virus. Clin. Infect. Dis. 2016, 63, 1584–1590. [Google Scholar] [CrossRef] [Green Version]
- Teo, T.H.; Her, Z.; Tan, J.J.; Lum, F.M.; Lee, W.W.; Chan, Y.H.; Ong, R.Y.; Kam, Y.W.; Leparc-Goffart, I.; Gallian, P.; et al. Caribbean and La Reunion chikungunya virus isolates differ in their capacity to induce proinflammatory Th1 and NK cell responses and acute joint pathology. J. Virol. 2015, 89, 7955–7969. [Google Scholar] [CrossRef]
- Wei Chiam, C.; Fun Chan, Y.; Chai Ong, K.; Thong Wong, K.; Sam, I.C. Neurovirulence comparison of chikungunya virus isolates of the Asian and East/Central/South African genotypes from Malaysia. J. Gen. Virol. 2015, 96, 3243–3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langsjoen, R.M.; Haller, S.L.; Roy, C.J.; Vinet-Oliphant, H.; Bergren, N.A.; Erasmus, J.H.; Livengood, J.A.; Powell, T.D.; Weaver, S.C.; Rossi, S.L. Chikungunya virus strains show lineage-specific variations in virulence and cross-protective ability in murine and nonhuman primate models. mBio 2018, 9, e02449-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messaoudi, I.; Vomaske, J.; Totonchy, T.; Kreklywich, C.N.; Haberthur, K.; Springgay, L.; Brien, J.D.; Diamond, M.S.; Defilippis, V.R.; Streblow, D.N. Chikungunya virus infection results in higher and persistent viral replication in aged rhesus macaques due to defects in anti-viral immunity. PLoS Negl. Trop. Dis. 2013, 7, e2343. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.K.; Tripathi, A. Association of toll-like receptor polymorphisms with susceptibility to chikungunya virus infection. Virology 2017, 511, 207–213. [Google Scholar] [CrossRef]
- Sengupta, S.; Mukherjee, S.; Bhattacharya, N.; Tripathi, A. Differential genotypic signatures of Toll-like receptor polymorphisms among dengue-chikungunya mono- and co-infected Eastern Indian patients. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Chaaithanya, I.K.; Muruganandam, N.; Surya, P.; Anwesh, M.; Alagarasu, K.; Vijayachari, P. Association of oligoadenylate synthetase gene cluster and DC-SIGN (CD209) gene polymorphisms with clinical symptoms in chikungunya virus infection. DNA Cell Biol. 2016, 35, 44–50. [Google Scholar] [CrossRef]
- Bucardo, F.; Reyes, Y.; Morales, M.; Briceno, R.; Gonzalez, F.; Lundkvist, A.; Svensson, L.; Nordgren, J. Association of genetic polymorphisms in DC-SIGN, Toll-like receptor 3, and Tumor Necrosis Factor α genes and the Lewis-negative phenotype with chikungunya infection and disease in Nicaragua. J. Infect. Dis. 2021, 223, 278–286. [Google Scholar] [CrossRef]
- Her, Z.; Teng, T.S.; Tan, J.J.; Teo, T.H.; Kam, Y.W.; Lum, F.M.; Lee, W.W.; Gabriel, C.; Melchiotti, R.; Andiappan, A.K.; et al. Loss of TLR3 aggravates CHIKV replication and pathology due to an altered virus-specific neutralizing antibody response. EMBO Mol. Med. 2015, 7, 24–41. [Google Scholar] [CrossRef]
- Ferreira, J.M.; Santos, L.D.S.; Oliveira, S.P.; Dos Santos, B.R.C.; Dos Santos, A.C.M.; de Moura, E.L.; de Souza, E.V.M.; de Lima Filho, J.L. Chikungunya virus infection outcome: A systematic review of host genetics. Immunol. Invest. 2021, 50, 58–79. [Google Scholar] [CrossRef]
- Dupuis-Maguiraga, L.; Noret, M.; Brun, S.; Le Grand, R.; Gras, G.; Roques, P. Chikungunya disease: Infection-associated markers from the acute to the chronic phase of arbovirus-induced arthralgia. PLoS Negl. Trop. Dis. 2012, 6, e1446. [Google Scholar] [CrossRef]
- Wauquier, N.; Becquart, P.; Nkoghe, D.; Padilla, C.; Ndjoyi-Mbiguino, A.; Leroy, E.M. The acute phase of chikungunya virus infection in humans is associated with strong innate immunity and T CD8 cell activation. J. Infect. Dis. 2011, 204, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Hakim, M.S.; Ikram, A.; Zhou, J.; Wang, W.; Peppelenbosch, M.P.; Pan, Q. Immunity against hepatitis E virus infection: Implications for therapy and vaccine development. Rev. Med. Virol. 2018, 28, e1964. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.E.; Locke, M.C.; Young, A.R.; Monte, K.; Hedberg, M.L.; Shimak, R.M.; Sheehan, K.C.F.; Veis, D.J.; Diamond, M.S.; Lenschow, D.J. Distinct roles of interferon alpha and beta in controlling chikungunya virus replication and modulating neutrophil-mediated inflammation. J. Virol. 2019, 94, e00841-19. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.A.; Wilson, J.; Gardner, J.; Larcher, T.; Babarit, C.; Le, T.T.; Anraku, I.; Kumagai, Y.; Loo, Y.M.; Gale, M., Jr.; et al. Interferon response factors 3 and 7 protect against chikungunya virus hemorrhagic fever and shock. J. Virol. 2012, 86, 9888–9898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webster, B.; Werneke, S.W.; Zafirova, B.; This, S.; Coleon, S.; Decembre, E.; Paidassi, H.; Bouvier, I.; Joubert, P.E.; Duffy, D.; et al. Plasmacytoid dendritic cells control dengue and chikungunya virus infections via IRF7-regulated interferon responses. Elife 2018, 7, e34273. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Scholte, F.E.; Chiang, C.; Albulescu, I.C.; Nichols, C.; He, Z.; Lin, R.; Snijder, E.J.; van Hemert, M.J.; Hiscott, J. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J. Virol. 2014, 88, 4180–4194. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, I.S.B.; Santos, E.C.; Tanabe, E.L.L.; Souza, S.J.M.; Santos, F.E.F.; Taniele-Silva, J.; Ferro, J.F.G.; Lima, M.C.; Moura, A.A.; Anderson, L.; et al. Cytokines and chemokines triggered by chikungunya virus infection in human patients during the very early acute phase. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 730–733. [Google Scholar] [CrossRef]
- Gallegos, K.M.; Drusano, G.L.; DZ, D.A.; Brown, A.N. Chikungunya virus: In vitro response to combination therapy with ribavirin and interferon alfa 2a. J. Infect. Dis. 2016, 214, 1192–1197. [Google Scholar] [CrossRef] [Green Version]
- Franco, E.J.; Tao, X.; Hanrahan, K.C.; Zhou, J.; Bulitta, J.B.; Brown, A.N. Combination regimens of favipiravir plus interferon alpha inhibit chikungunya virus replication in clinically relevant human cell lines. Microorganisms 2021, 9, 307. [Google Scholar] [CrossRef]
- Locke, M.C.; Fox, L.E.; Dunlap, B.F.; Young, A.R.; Monte, K.; Lenschow, D.J. Interferon alpha, but not interferon beta, acts early to control chronic chikungunya virus pathogenesis. J. Virol. 2022, 96, e0114321. [Google Scholar] [CrossRef]
- Couderc, T.; Chretien, F.; Schilte, C.; Disson, O.; Brigitte, M.; Guivel-Benhassine, F.; Touret, Y.; Barau, G.; Cayet, N.; Schuffenecker, I.; et al. A mouse model for chikungunya: Young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 2008, 4, e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilte, C.; Couderc, T.; Chretien, F.; Sourisseau, M.; Gangneux, N.; Guivel-Benhassine, F.; Kraxner, A.; Tschopp, J.; Higgs, S.; Michault, A.; et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 2010, 207, 429–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Min, N.; Ma, L.; Mok, C.K.; Chu, J.J.H. Adenovirus vectored IFN-alpha protects mice from lethal challenge of chikungunya virus infection. PLoS Negl. Trop. Dis. 2020, 14, e0008910. [Google Scholar] [CrossRef]
- Colavita, F.; Vita, S.; Lalle, E.; Carletti, F.; Bordi, L.; Vincenti, D.; Pozzetto, I.; Aiuti, M.; Vairo, F.; Capobianchi, M.R.; et al. Overproduction of IL-6 and type-I IFN in a lethal case of chikungunya virus infection in an elderly man during the 2017 Italian outbreak. Open Forum Infect. Dis. 2018, 5, ofy276. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, M.; Lozhkov, A.; Romanovskaya-Romanko, E.; Baranovskaya, I.; Sergeeva, M.; Klotchenko, S.; Vasin, A. IFN-λ1 displays various levels of antiviral activity in vitro in a select panel of RNA viruses. Viruses 2021, 13, 1602. [Google Scholar] [CrossRef]
- Wichit, S.; Hamel, R.; Zanzoni, A.; Diop, F.; Cribier, A.; Talignani, L.; Diack, A.; Ferraris, P.; Liegeois, F.; Urbach, S.; et al. SAMHD1 enhances chikungunya and Zika virus replication in human skin fibroblasts. Int. J. Mol. Sci. 2019, 20, 1695. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.R.; Abraham, R.; Sundaram, S.; Sreekumar, E. Interferon regulated gene (IRG) expression-signature in a mouse model of chikungunya virus neurovirulence. J. Neurovirol. 2017, 23, 886–902. [Google Scholar] [CrossRef]
- Wichit, S.; Hamel, R.; Yainoy, S.; Gumpangseth, N.; Panich, S.; Phuadraksa, T.; Saetear, P.; Monteil, A.; Morales Vargas, R.; Misse, D. Interferon-inducible protein (IFI) 16 regulates chikungunya and Zika virus infection in human skin fibroblasts. EXCLI J. 2019, 18, 467–476. [Google Scholar] [CrossRef]
- Franz, S.; Pott, F.; Zillinger, T.; Schuler, C.; Dapa, S.; Fischer, C.; Passos, V.; Stenzel, S.; Chen, F.; Dohner, K.; et al. Human IFITM3 restricts chikungunya virus and Mayaro virus infection and is susceptible to virus-mediated counteraction. Life Sci. Alliance 2021, 4, e202000909. [Google Scholar] [CrossRef]
- Poddar, S.; Hyde, J.L.; Gorman, M.J.; Farzan, M.; Diamond, M.S. The interferon-stimulated gene IFITM3 restricts infection and pathogenesis of arthritogenic and encephalitic alphaviruses. J. Virol. 2016, 90, 8780–8794. [Google Scholar] [CrossRef]
- Taschuk, F.; Tapescu, I.; Moy, R.H.; Cherry, S. DDX56 binds to chikungunya virus RNA to control infection. mBio 2020, 11, e02623-20. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Poddar, S.; Shimak, R.M.; Diamond, M.S. Interferon regulatory factor 1 protects against chikungunya virus-induced immunopathology by restricting infection in muscle cells. J. Virol. 2017, 91, e01419-17. [Google Scholar] [CrossRef] [Green Version]
- Weiss, C.M.; Trobaugh, D.W.; Sun, C.; Lucas, T.M.; Diamond, M.S.; Ryman, K.D.; Klimstra, W.B. The interferon-induced exonuclease ISG20 exerts antiviral activity through upregulation of type I interferon response proteins. mSphere 2018, 3, e00209–e00218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, E.S.; Shrihari, S.; Hykes, B.L., Jr.; Handley, S.A.; Andhey, P.S.; Huang, Y.S.; Swain, A.; Droit, L.; Chebrolu, K.K.; Mack, M.; et al. The intestinal microbiome restricts Alphavirus infection and dissemination through a bile acid-type I IFN signaling axis. Cell 2020, 182, 901–918.e918. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Lee, J.Y.; Myoung, J. Chikungunya virus-encoded nsP2, E2 and E1 strongly antagonize the interferon-beta signaling pathway. J. Microbiol. Biotechnol. 2019, 29, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Lee, J.Y.; Myoung, J. Chikungunya virus nsP2 impairs MDA5/RIG-I-mediated induction of NF-kappaB promoter activation: A potential target for virus-specific therapeutics. J. Microbiol. Biotechnol. 2020, 30, 1801–1809. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Lopez, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Interleukin 27 as an inducer of antiviral response against chikungunya virus infection in human macrophages. Cell Immunol. 2021, 367, 104411. [Google Scholar] [CrossRef]
- Fros, J.J.; Liu, W.J.; Prow, N.A.; Geertsema, C.; Ligtenberg, M.; Vanlandingham, D.L.; Schnettler, E.; Vlak, J.M.; Suhrbier, A.; Khromykh, A.A.; et al. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J. Virol. 2010, 84, 10877–10887. [Google Scholar] [CrossRef] [Green Version]
- Fros, J.J.; van der Maten, E.; Vlak, J.M.; Pijlman, G.P. The C-terminal domain of chikungunya virus nsP2 independently governs viral RNA replication, cytopathicity, and inhibition of interferon signaling. J. Virol. 2013, 87, 10394–10400. [Google Scholar] [CrossRef] [Green Version]
- Goertz, G.P.; McNally, K.L.; Robertson, S.J.; Best, S.M.; Pijlman, G.P.; Fros, J.J. The methyltransferase-like domain of chikungunya virus nsP2 inhibits the interferon response by promoting the nuclear export of STAT1. J. Virol. 2018, 92, e01008-18. [Google Scholar] [CrossRef]
- Wichit, S.; Diop, F.; Hamel, R.; Talignani, L.; Ferraris, P.; Cornelie, S.; Liegeois, F.; Thomas, F.; Yssel, H.; Misse, D. Aedes aegypti saliva enhances chikungunya virus replication in human skin fibroblasts via inhibition of the type I interferon signaling pathway. Infect. Genet. Evol. 2017, 55, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Lum, F.M.; Teo, T.H.; Lee, W.W.; Kam, Y.W.; Renia, L.; Ng, L.F. An essential role of antibodies in the control of chikungunya virus infection. J. Immunol. 2013, 190, 6295–6302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoarau, J.J.; Gay, F.; Pelle, O.; Samri, A.; Jaffar-Bandjee, M.C.; Gasque, P.; Autran, B. Identical strength of the T cell responses against E2, nsP1 and capsid CHIKV proteins in recovered and chronic patients after the epidemics of 2005–2006 in La Reunion Island. PLoS ONE 2013, 8, e84695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, T.H.; Lum, F.M.; Claser, C.; Lulla, V.; Lulla, A.; Merits, A.; Renia, L.; Ng, L.F. A pathogenic role for CD4+ T cells during chikungunya virus infection in mice. J. Immunol. 2013, 190, 259–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, C.N.S.; Gois, B.M.; Lima, V.S.; Guerra-Gomes, I.C.; Araujo, J.M.G.; Gomes, J.A.S.; Araujo, D.A.M.; Medeiros, I.A.; Azevedo, F.; Veras, R.C.; et al. Human CD8 T-cell activation in acute and chronic chikungunya infection. Immunology 2018, 155, 499–504. [Google Scholar] [CrossRef]
- Davenport, B.J.; Bullock, C.; McCarthy, M.K.; Hawman, D.W.; Murphy, K.M.; Kedl, R.M.; Diamond, M.S.; Morrison, T.E. Chikungunya virus evades antiviral CD8(+) T cell responses to establish persistent infection in joint-associated tissues. J. Virol. 2020, 94, e02036-19. [Google Scholar] [CrossRef]
- Kulkarni, S.P.; Ganu, M.; Jayawant, P.; Thanapati, S.; Ganu, A.; Tripathy, A.S. Regulatory T cells and IL-10 as modulators of chikungunya disease outcome: A preliminary study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2475–2481. [Google Scholar] [CrossRef]
- Gois, B.M.; Peixoto, R.F.; Guerra-Gomes, I.C.; Palmeira, P.H.S.; Dias, C.N.S.; Araujo, J.M.G.; Veras, R.C.; Medeiros, I.A.; Azevedo, F.; Boyton, R.J.; et al. Regulatory T cells in acute and chronic human chikungunya infection. Microbes Infect. 2022, 24, 104927. [Google Scholar] [CrossRef]
- Lee, W.W.; Teo, T.H.; Her, Z.; Lum, F.M.; Kam, Y.W.; Haase, D.; Renia, L.; Rotzschke, O.; Ng, L.F. Expanding regulatory T cells alleviates chikungunya virus-induced pathology in mice. J. Virol. 2015, 89, 7893–7904. [Google Scholar] [CrossRef] [Green Version]
- Venugopalan, A.; Ghorpade, R.P.; Chopra, A. Cytokines in acute chikungunya. PLoS ONE 2014, 9, e111305. [Google Scholar] [CrossRef]
- Chelluboina, S.; Robin, S.; Aswathyraj, S.; Arunkumar, G. Persistence of antibody response in chikungunya. Virusdisease 2019, 30, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Jain, J.; Okabayashi, T.; Kaur, N.; Nakayama, E.; Shioda, T.; Gaind, R.; Kurosu, T.; Sunil, S. Evaluation of an immunochromatography rapid diagnosis kit for detection of chikungunya virus antigen in India, a dengue-endemic country. Virol. J. 2018, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Chua, C.L.; Sam, I.C.; Chiam, C.W.; Chan, Y.F. The neutralizing role of IgM during early chikungunya virus infection. PLoS ONE 2017, 12, e0171989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prince, H.E.; Seaton, B.L.; Matud, J.L.; Batterman, H.J. Chikungunya virus RNA and antibody testing at a National Reference Laboratory since the emergence of chikungunya virus in the Americas. Clin. Vaccine Immunol. 2015, 22, 291–297. [Google Scholar] [CrossRef] [Green Version]
- Porter, K.R.; Tan, R.; Istary, Y.; Suharyono, W.; Sutaryo; Widjaja, S.; Ma’Roef, C.; Listiyaningsih, E.; Kosasih, H.; Hueston, L.; et al. A serological study of chikungunya virus transmission in Yogyakarta, Indonesia: Evidence for the first outbreak since 1982. Southeast Asian J. Trop. Med. Public Health 2004, 35, 408–415. [Google Scholar]
- Quiroz, J.A.; Malonis, R.J.; Thackray, L.B.; Cohen, C.A.; Pallesen, J.; Jangra, R.K.; Brown, R.S.; Hofmann, D.; Holtsberg, F.W.; Shulenin, S.; et al. Human monoclonal antibodies against chikungunya virus target multiple distinct epitopes in the E1 and E2 glycoproteins. PLoS Pathog. 2019, 15, e1008061. [Google Scholar] [CrossRef] [Green Version]
- Kose, N.; Fox, J.M.; Sapparapu, G.; Bombardi, R.; Tennekoon, R.N.; de Silva, A.D.; Elbashir, S.M.; Theisen, M.A.; Humphris-Narayanan, E.; Ciaramella, G.; et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019, 4, eaaw6647. [Google Scholar] [CrossRef]
- Broeckel, R.; Fox, J.M.; Haese, N.; Kreklywich, C.N.; Sukulpovi-Petty, S.; Legasse, A.; Smith, P.P.; Denton, M.; Corvey, C.; Krishnan, S.; et al. Therapeutic administration of a recombinant human monoclonal antibody reduces the severity of chikungunya virus disease in rhesus macaques. PLoS Negl. Trop. Dis. 2017, 11, e0005637. [Google Scholar] [CrossRef] [Green Version]
- Anfasa, F.; Lim, S.M.; Fekken, S.; Wever, R.; Osterhaus, A.; Martina, B.E.E. Characterization of antibody response in patients with acute and chronic chikungunya virus disease. J. Clin. Virol. 2019, 117, 68–72. [Google Scholar] [CrossRef]
- Kam, Y.W.; Simarmata, D.; Chow, A.; Her, Z.; Teng, T.S.; Ong, E.K.; Renia, L.; Leo, Y.S.; Ng, L.F. Early appearance of neutralizing immunoglobulin G3 antibodies is associated with chikungunya virus clearance and long-term clinical protection. J. Infect. Dis. 2012, 205, 1147–1154. [Google Scholar] [CrossRef]
- Henss, L.; Yue, C.; Von Rhein, C.; Tschismarov, R.; Lewis-Ximenez, L.L.; Dolle, A.; Baylis, S.A.; Schnierle, B.S. Analysis of humoral immune responses in chikungunya virus (CHIKV)-infected patients and individuals vaccinated with a candidate CHIKV vaccine. J. Infect. Dis. 2020, 221, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Kam, Y.W.; Lum, F.M.; Teo, T.H.; Lee, W.W.; Simarmata, D.; Harjanto, S.; Chua, C.L.; Chan, Y.F.; Wee, J.K.; Chow, A.; et al. Early neutralizing IgG response to chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol. Med. 2012, 4, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Tumkosit, U.; Siripanyaphinyo, U.; Takeda, N.; Tsuji, M.; Maeda, Y.; Ruchusatsawat, K.; Shioda, T.; Mizushima, H.; Chetanachan, P.; Wongjaroen, P.; et al. Anti-chikungunya virus monoclonal antibody that inhibits viral fusion and release. J. Virol. 2020, 94, e00252-20. [Google Scholar] [CrossRef] [PubMed]
- Lum, F.M.; Couderc, T.; Chia, B.S.; Ong, R.Y.; Her, Z.; Chow, A.; Leo, Y.S.; Kam, Y.W.; Renia, L.; Lecuit, M.; et al. Antibody-mediated enhancement aggravates chikungunya virus infection and disease severity. Sci. Rep. 2018, 8, 1860. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.; Desai, A.; Krishna, S.S.; Vasanthapuram, R. Molecular mimicry between chikungunya virus and host components: A possible mechanism for the arthritic manifestations. PLoS Negl. Trop. Dis. 2017, 11, e0005238. [Google Scholar] [CrossRef]
- De Carvalho, J.F.; Kanduc, D.; da Silva, F.F.; Tanay, A.; Lucchese, A.; Shoenfeld, Y. Sjogren’s syndrome associated with chikungunya infection: A case report. Rheumatol. Ther. 2021, 8, 631–637. [Google Scholar] [CrossRef]
- Kuno, G. A re-examination of the history of etiologic confusion between dengue and chikungunya. PLoS Negl. Trop. Dis. 2015, 9, e0004101. [Google Scholar] [CrossRef] [Green Version]
- Simon, F.; Javelle, E.; Cabie, A.; Bouquillard, E.; Troisgros, O.; Gentile, G.; Leparc-Goffart, I.; Hoen, B.; Gandjbakhch, F.; Rene-Corail, P.; et al. French guidelines for the management of chikungunya (acute and persistent presentations). November 2014. Med. Mal. Infect. 2015, 45, 243–263. [Google Scholar] [CrossRef]
- Staikowsky, F.; Talarmin, F.; Grivard, P.; Souab, A.; Schuffenecker, I.; Le Roux, K.; Lecuit, M.; Michault, A. Prospective study of chikungunya virus acute infection in the Island of La Reunion during the 2005-2006 outbreak. PLoS ONE 2009, 4, e7603. [Google Scholar] [CrossRef] [Green Version]
- Gay, N.; Rousset, D.; Huc, P.; Matheus, S.; Ledrans, M.; Rosine, J.; Cassadou, S.; Noel, H. Seroprevalence of Asian lineage chikungunya virus infection on Saint Martin Island, 7 months after the 2013 emergence. Am. J. Trop. Med. Hyg. 2016, 94, 393–396. [Google Scholar] [CrossRef]
- Rudolph, K.E.; Lessler, J.; Moloney, R.M.; Kmush, B.; Cummings, D.A. Incubation periods of mosquito-borne viral infections: A systematic review. Am. J. Trop. Med. Hyg. 2014, 90, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.D.; Valverde, J.G.; Morais, I.C.; Souza, C.R.M.; Fagundes Neto, J.C.; Melo, M.F.; Nascimento, Y.M.; Alves, B.E.B.; Medeiros, L.G.; Pereira, H.W.B.; et al. Epidemiologic and clinical investigations during a chikungunya outbreak in Rio Grande do Norte State, Brazil. PLoS ONE 2020, 15, e0241799. [Google Scholar] [CrossRef]
- Hossain, M.S.; Hasan, M.M.; Islam, M.S.; Islam, S.; Mozaffor, M.; Khan, M.A.S.; Ahmed, N.; Akhtar, W.; Chowdhury, S.; Arafat, S.M.Y.; et al. Chikungunya outbreak (2017) in Bangladesh: Clinical profile, economic impact and quality of life during the acute phase of the disease. PLoS Negl. Trop. Dis. 2018, 12, e0006561. [Google Scholar] [CrossRef] [PubMed]
- Imad, H.A.; Phadungsombat, J.; Nakayama, E.E.; Suzuki, K.; Ibrahim, A.M.; Afaa, A.; Azeema, A.; Nazfa, A.; Yazfa, A.; Ahmed, A.; et al. Clinical features of acute chikungunya virus infection in children and adults during an outbreak in the Maldives. Am. J. Trop. Med. Hyg. 2021, 105, 946–954. [Google Scholar] [CrossRef]
- Borgherini, G.; Poubeau, P.; Staikowsky, F.; Lory, M.; Le Moullec, N.; Becquart, J.P.; Wengling, C.; Michault, A.; Paganin, F. Outbreak of chikungunya on Reunion Island: Early clinical and laboratory features in 157 adult patients. Clin. Infect. Dis. 2007, 44, 1401–1407. [Google Scholar] [CrossRef] [Green Version]
- Simon, F.; Parola, P.; Grandadam, M.; Fourcade, S.; Oliver, M.; Brouqui, P.; Hance, P.; Kraemer, P.; Mohamed, A.A.; de Lamballerie, X.; et al. Chikungunya infection: An emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine 2007, 86, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Zaid, A.; Gerardin, P.; Taylor, A.; Mostafavi, H.; Malvy, D.; Mahalingam, S. Chikungunya arthritis: Implications of acute and chronic inflammation mechanisms on disease management. Arthritis Rheumatol. 2018, 70, 484–495. [Google Scholar] [CrossRef] [Green Version]
- Javelle, E.; Tiong, T.H.; Leparc-Goffart, I.; Savini, H.; Simon, F. Inflammation of the external ear in acute chikungunya infection: Experience from the outbreak in Johor Bahru, Malaysia, 2008. J. Clin. Virol. 2014, 59, 270–273. [Google Scholar] [CrossRef]
- Blettery, M.; Brunier, L.; Polomat, K.; Moinet, F.; Deligny, C.; Arfi, S.; Jean-Baptiste, G.; De Bandt, M. Brief report: Management of chronic post-chikungunya rheumatic disease: The Martinican experience. Arthritis Rheumatol. 2016, 68, 2817–2824. [Google Scholar] [CrossRef]
- Godaert, L.; Najioullah, F.; Bartholet, S.; Colas, S.; Yactayo, S.; Cabie, A.; Fanon, J.L.; Cesaire, R.; Drame, M. Atypical clinical presentations of acute phase chikungunya virus infection in older adults. J. Am. Geriatr. Soc. 2017, 65, 2510–2515. [Google Scholar] [CrossRef]
- Economopoulou, A.; Dominguez, M.; Helynck, B.; Sissoko, D.; Wichmann, O.; Quenel, P.; Germonneau, P.; Quatresous, I. Atypical chikungunya virus infections: Clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Reunion. Epidemiol. Infect. 2009, 137, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Tandale, B.V.; Sathe, P.S.; Arankalle, V.A.; Wadia, R.S.; Kulkarni, R.; Shah, S.V.; Shah, S.K.; Sheth, J.K.; Sudeep, A.B.; Tripathy, A.S.; et al. Systemic involvements and fatalities during chikungunya epidemic in India, 2006. J. Clin. Virol. 2009, 46, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.R.; Leopoldo Codova, G.; Castro, J.S.; Rodriguez, L.; Saravia, V.; Arvelaez, J.; Rios-Fabra, A.; Longhi, M.A.; Marcano, M. Chikungunya fever: Atypical and lethal cases in the Western hemisphere: A Venezuelan experience. IDCases 2015, 2, 6–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, R.; Gerardin, P.; de Brito, C.A.A.; Soares, C.N.; Ferreira, M.L.B.; Solomon, T. The neurological complications of chikungunya virus: A systematic review. Rev. Med. Virol. 2018, 28, e1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, R.; Viana, R.; Brainer-Lima, A.; FloreAncio, T.; Carvalho, M.D.; van der Linden, V.; Amorim, A.; Rocha, M.A.; Medeiros, F. Perinatal chikungunya virus-associated encephalitis leading to postnatal-onset microcephaly and optic atrophy. Pediatr. Infect. Dis. J. 2018, 37, 94–95. [Google Scholar] [CrossRef] [PubMed]
- Villamil-Gomez, W.; Alba-Silvera, L.; Menco-Ramos, A.; Gonzalez-Vergara, A.; Molinares-Palacios, T.; Barrios-Corrales, M.; Rodriguez-Morales, A.J. Congenital chikungunya virus infection in Sincelejo, Colombia: A case series. J. Trop. Pediatr. 2015, 61, 386–392. [Google Scholar] [CrossRef] [Green Version]
- Nyamwaya, D.K.; Otiende, M.; Omuoyo, D.O.; Githinji, G.; Karanja, H.K.; Gitonga, J.N.; de Laurent, Z.R.; Otieno, J.R.; Sang, R.; Kamau, E.; et al. Endemic chikungunya fever in Kenyan children: A prospective cohort study. BMC Infect. Dis. 2021, 21, 186. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Her, Z.; Ong, E.K.; Chen, J.M.; Dimatatac, F.; Kwek, D.J.; Barkham, T.; Yang, H.; Renia, L.; Leo, Y.S.; et al. Persistent arthralgia induced by chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J. Infect. Dis. 2011, 203, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Hoarau, J.J.; Jaffar Bandjee, M.C.; Krejbich Trotot, P.; Das, T.; Li-Pat-Yuen, G.; Dassa, B.; Denizot, M.; Guichard, E.; Ribera, A.; Henni, T.; et al. Persistent chronic inflammation and infection by chikungunya arthritogenic alphavirus in spite of a robust host immune response. J. Immunol. 2010, 184, 5914–5927. [Google Scholar] [CrossRef] [Green Version]
- Mohan, A.; Kiran, D.H.; Manohar, I.C.; Kumar, D.P. Epidemiology, clinical manifestations, and diagnosis of chikungunya fever: Lessons learned from the re-emerging epidemic. Indian J. Dermatol. 2010, 55, 54–63. [Google Scholar] [CrossRef]
- Simon, F.; Javelle, E.; Oliver, M.; Leparc-Goffart, I.; Marimoutou, C. Chikungunya virus infection. Curr. Infect. Dis. Rep. 2011, 13, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti, A.; Thioune, M.; Abel, S.; Belrose, G.; Calmont, I.; Cesaire, R.; Cervantes, M.; Fagour, L.; Javelle, E.; Lebris, C.; et al. Prevalence of chronic chikungunya and associated risks factors in the French West Indies (La Martinique): A prospective cohort study. PLoS Negl. Trop. Dis. 2020, 14, e0007327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention. Chikungunya Virus: Clinical Evaluation & Disease. Available online: https://www.cdc.gov/chikungunya/hc/clinicalevaluation.html (accessed on 22 April 2022).
- Sissoko, D.; Ezzedine, K.; Moendandze, A.; Giry, C.; Renault, P.; Malvy, D. Field evaluation of clinical features during chikungunya outbreak in Mayotte, 2005–2006. Trop. Med. Int. Health 2010, 15, 600–607. [Google Scholar] [CrossRef]
- Johnson, B.W.; Russell, B.J.; Goodman, C.H. Laboratory diagnosis of chikungunya virus infections and commercial sources for diagnostic assays. J. Infect. Dis. 2016, 214, S471–S474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patramool, S.; Bernard, E.; Hamel, R.; Natthanej, L.; Chazal, N.; Surasombatpattana, P.; Ekchariyawat, P.; Daoust, S.; Thongrungkiat, S.; Thomas, F.; et al. Isolation of infectious chikungunya virus and dengue virus using anionic polymer-coated magnetic beads. J. Virol. Methods 2013, 193, 55–61. [Google Scholar] [CrossRef]
- Cunha, R.V.D.; Trinta, K.S. Chikungunya virus: Clinical aspects and treatment—A review. Memórias Inst. Oswaldo Cruz 2017, 112, 523–531. [Google Scholar] [CrossRef]
- Riswari, S.F.; Ma’roef, C.N.; Djauhari, H.; Kosasih, H.; Perkasa, A.; Yudhaputri, F.A.; Artika, I.M.; Williams, M.; van der Ven, A.; Myint, K.S.; et al. Study of viremic profile in febrile specimens of chikungunya in Bandung, Indonesia. J. Clin. Virol. 2016, 74, 61–65. [Google Scholar] [CrossRef] [Green Version]
- Wikan, N.; Sakoonwatanyoo, P.; Ubol, S.; Yoksan, S.; Smith, D.R. Chikungunya virus infection of cell lines: Analysis of the East, Central and South African lineage. PLoS ONE 2012, 7, e31102. [Google Scholar] [CrossRef] [Green Version]
- Sudeep, A.B.; Vyas, P.B.; Parashar, D.; Shil, P. Differential susceptibility & replication potential of Vero E6, BHK-21, RD, A-549, C6/36 cells & Aedes aegypti mosquitoes to three strains of chikungunya virus. Indian J. Med. Res. 2019, 149, 771–777. [Google Scholar] [CrossRef]
- Pyndiah, M.N.; Pursem, V.; Meetoo, G.; Daby, S.; Ramuth, V.; Bhinkah, P.; Chuttoo, R.; Paratian, U. Chikungunya virus isolation using simplified cell culture technique in Mauritius. Med. Trop. 2012, 72, 63–65. [Google Scholar]
- Chiam, C.W.; Chan, Y.F.; Loong, S.K.; Yong, S.S.; Hooi, P.S.; Sam, I.C. Real-time polymerase chain reaction for diagnosis and quantitation of negative strand of chikungunya virus. Diagn. Microbiol. Infect. Dis. 2013, 77, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Broeders, S.; Garlant, L.; Fraiture, M.A.; Vandermassen, E.; Suin, V.; Vanhomwegen, J.; Dupont-Rouzeyrol, M.; Rousset, D.; Van Gucht, S.; Roosens, N. A new multiplex RT-qPCR method for the simultaneous detection and discrimination of Zika and chikungunya viruses. Int. J. Infect. Dis. 2020, 92, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.; Orba, Y.; Sequeira, P.C.; Sugimoto, C.; Hall, W.W.; Eshita, Y.; Suzuki, Y.; Runtuwene, L.; Brasil, P.; Calvet, G.; et al. Field diagnosis and genotyping of chikungunya virus using a dried reverse transcription loop-mediated isothermal amplification (LAMP) assay and MinION sequencing. PLoS Negl. Trop. Dis. 2019, 13, e0007480. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Jimena, B.; Wehner, S.; Harold, G.; Bakheit, M.; Frischmann, S.; Bekaert, M.; Faye, O.; Sall, A.A.; Weidmann, M. Development of a single-tube one-step RT-LAMP assay to detect the chikungunya virus genome. PLoS Negl. Trop. Dis. 2018, 12, e0006448. [Google Scholar] [CrossRef]
- Appassakij, H.; Khuntikij, P.; Kemapunmanus, M.; Wutthanarungsan, R.; Silpapojakul, K. Viremic profiles in asymptomatic and symptomatic chikungunya fever: A blood transfusion threat? Transfusion 2013, 53, 2567–2574. [Google Scholar] [CrossRef]
- Edwards, T.; Del Carmen Castillo Signor, L.; Williams, C.; Larcher, C.; Espinel, M.; Theaker, J.; Donis, E.; Cuevas, L.E.; Adams, E.R. Analytical and clinical performance of a chikungunya qRT-PCR for Central and South America. Diagn. Microbiol. Infect. Dis. 2017, 89, 35–39. [Google Scholar] [CrossRef]
- Thirion, L.; Pezzi, L.; Corcostegui, I.; Dubot-Peres, A.; Falchi, A.; de Lamballerie, X.; Charrel, R.N. Development and evaluation of a duo chikungunya virus real-time RT-PCR assay targeting two regions within the genome. Viruses 2019, 11, 755. [Google Scholar] [CrossRef] [Green Version]
- Giry, C.; Roquebert, B.; Li-Pat-Yuen, G.; Gasque, P.; Jaffar-Bandjee, M.C. Improved detection of genus-specific Alphavirus using a generic TaqMan(R) assay. BMC Microbiol. 2017, 17, 164. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.A.; Blitvich, B.J.; Johansen, C.A.; Blacksell, S.D. Advances in arbovirus surveillance, detection and diagnosis. J. Biomed. Biotechnol. 2012, 2012, 512969. [Google Scholar] [CrossRef] [Green Version]
- Martins, E.B.; Silva, M.F.B.; Tassinari, W.S.; de Bruycker-Nogueira, F.; Moraes, I.C.V.; Rodrigues, C.D.S.; Santos, C.C.; Sampaio, S.A.; Pina-Costa, A.; Fabri, A.A.; et al. Detection of chikungunya virus in bodily fluids: The INOVACHIK cohort study. PLoS Negl. Trop. Dis. 2022, 16, e0010242. [Google Scholar] [CrossRef]
- Manzoor, K.N.; Javed, F.; Ejaz, M.; Ali, M.; Mujaddadi, N.; Khan, A.A.; Khattak, A.A.; Zaib, A.; Ahmad, I.; Saeed, W.K.; et al. The global emergence of chikungunya infection: An integrated view. Rev. Med. Virol. 2021, 32, e2287. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Regional Office for South-East Asia. Guidelines for Prevention and Control of Chikungunya Fever. WHO Regional Office for South-East Asia: Geneva, Switzerland, 2009. Available online: https://apps.who.int/iris/handle/10665/205166 (accessed on 22 April 2022).
- Andrew, A.; Navien, T.N.; Yeoh, T.S.; Citartan, M.; Mangantig, E.; Sum, M.S.H.; Ch’ng, E.S.; Tang, T.H. Diagnostic accuracy of serological tests for the diagnosis of chikungunya virus infection: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2022, 16, e0010152. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Lopez-Camacho, C.; Garcia-Larragoiti, N.; Cano-Mendez, A.; Hernandez-Flores, K.G.; Dominguez-Aleman, C.A.; Mar, M.A.; Vivanco-Cid, H.; Viveros-Sandoval, M.E.; Reyes-Sandoval, A. Development of an E2 ELISA methodology to assess chikungunya seroprevalence in patients from an endemic region of Mexico. Viruses 2019, 11, 407. [Google Scholar] [CrossRef] [Green Version]
- Bagno, F.F.; Godoi, L.C.; Figueiredo, M.M.; Sergio, S.A.R.; Moraes, T.F.S.; Salazar, N.C.; Kim, Y.C.; Reyes-Sandoval, A.; da Fonseca, F.G. Chikungunya E2 protein produced in E. coli and HEK293-T cells-comparison of their performances in ELISA. Viruses 2020, 12, 939. [Google Scholar] [CrossRef]
- Powers, A.M.; Brault, A.C.; Tesh, R.B.; Weaver, S.C. Re-emergence of chikungunya and O’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000, 81, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Clements, T.L.; Rossi, C.A.; Irish, A.K.; Kibuuka, H.; Eller, L.A.; Robb, M.L.; Kataaha, P.; Michael, N.L.; Hensley, L.E.; Schoepp, R.J. Chikungunya and O’nyong-nyong viruses in Uganda: Implications for diagnostics. Open Forum Infect. Dis. 2019, 6, ofz001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, C.; Bozza, F.; Merino Merino, X.J.; Pedroso, C.; de Oliveira Filho, E.F.; Moreira-Soto, A.; Schwalb, A.; de Lamballerie, X.; Netto, E.M.; Bozza, P.T.; et al. Robustness of serologic investigations for chikungunya and Mayaro viruses following coemergence. mSphere 2020, 5, e00915–e00919. [Google Scholar] [CrossRef] [Green Version]
- Lima, M.; de Lima, R.C.; de Azeredo, E.L.; Dos Santos, F.B. Analysis of a routinely used commercial anti-chikungunya IgM ELISA reveals cross-reactivities with dengue in Brazil: A new challenge for differential diagnosis? Diagnostics 2021, 11, 819. [Google Scholar] [CrossRef]
- Fritzell, C.; Rousset, D.; Adde, A.; Kazanji, M.; Van Kerkhove, M.D.; Flamand, C. Current challenges and implications for dengue, chikungunya and zika seroprevalence studies worldwide: A scoping review. PLoS Negl. Trop. Dis. 2018, 12, e0006533. [Google Scholar] [CrossRef] [Green Version]
- Huits, R.; Okabayashi, T.; Cnops, L.; Barbe, B.; Van Den Berg, R.; Bartholomeeusen, K.; Arien, K.K.; Jacobs, J.; Bottieau, E.; Nakayama, E.E.; et al. Diagnostic accuracy of a rapid E1-antigen test for chikungunya virus infection in a reference setting. Clin. Microbiol. Infect. 2018, 24, 78–81. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.; Bosch, I.; Salcedo, N.; Herrera, B.B.; de Puig, H.; Narvaez, C.F.; Caicedo-Borrero, D.M.; Lorenzana, I.; Parham, L.; Garcia, K.; et al. Development and validation of a rapid lateral flow E1/E2-antigen test and ELISA in patients infected with emerging Asian strain of chikungunya virus in the Americas. Viruses 2020, 12, 971. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Dhanwani, R.; Kumar, J.S.; Rao, P.V.; Parida, M. Comparative evaluation of the diagnostic potential of recombinant envelope proteins and native cell culture purified viral antigens of chikungunya virus. J. Med. Virol. 2014, 86, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Huits, R.; Phadungsombat, J.; Tuekprakhon, A.; Nakayama, E.E.; van den Berg, R.; Barbe, B.; Cnops, L.; Rahim, R.; Hasan, A.; et al. Promising application of monoclonal antibody against chikungunya virus E1-antigen across genotypes in immunochromatographic rapid diagnostic tests. Virol. J. 2020, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.C.; Park, H.; Shin, H.J.; Nguyen, N.M.; Nguyen, A.T.V.; Trinh, T.T.; Duong, T.H.Y.; Tuong, H.T.; Hoang, V.T.; Seo, G.E.; et al. Fluorescent immunosorbent assay for chikungunya virus detection. Intervirology 2019, 62, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Tuekprakhon, A.; Nakayama, E.E.; Bartholomeeusen, K.; Puiprom, O.; Sasaki, T.; Huits, R.; Luplertlop, N.; Kosoltanapiwat, N.; Maneekan, P.; Arien, K.K.; et al. Variation at position 350 in the chikungunya virus 6K-E1 protein determines the sensitivity of detection in a rapid E1-antigen test. Sci. Rep. 2018, 8, 1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Song, S.; Zhang, L. Recent progress in vaccine development against chikungunya virus. Front. Microbiol. 2019, 10, 2881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.; Sudeep, A.B.; Arankalle, V.A. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine 2012, 30, 6142–6149. [Google Scholar] [CrossRef]
- Amaral, M.P.; Coirada, F.C.; de Souza Apostolico, J.; Tomita, N.; Fernandes, E.R.; Santos Souza, H.F.; Chura-Chambi, R.M.; Morganti, L.; Boscardin, S.B.; Rosa, D.S. Prime-boost with chikungunya virus E2 envelope protein combined with Poly (I:C) induces specific humoral and cellular immune responses. Curr. Res. Immunol. 2021, 2, 23–31. [Google Scholar] [CrossRef]
- Khan, M.; Dhanwani, R.; Rao, P.V.; Parida, M. Subunit vaccine formulations based on recombinant envelope proteins of chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus Res. 2012, 167, 236–246. [Google Scholar] [CrossRef]
- Chen, G.L.; Coates, E.E.; Plummer, S.H.; Carter, C.A.; Berkowitz, N.; Conan-Cibotti, M.; Cox, J.H.; Beck, A.; O’Callahan, M.; Andrews, C.; et al. Effect of a chikungunya virus-like particle vaccine on safety and tolerability outcomes: A randomized clinical trial. JAMA 2020, 323, 1369–1377. [Google Scholar] [CrossRef]
- Chang, L.J.; Dowd, K.A.; Mendoza, F.H.; Saunders, J.G.; Sitar, S.; Plummer, S.H.; Yamshchikov, G.; Sarwar, U.N.; Hu, Z.; Enama, M.E.; et al. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: A phase 1 dose-escalation trial. Lancet 2014, 384, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Bennett, S.R.; McCarty, J.M.; Ramanathan, R.; Mendy, J.; Richardson, J.S.; Smith, J.; Alexander, J.; Ledgerwood, J.E.; de Lame, P.A.; Royalty Tredo, S.; et al. Safety and immunogenicity of PXVX0317, an aluminium hydroxide-adjuvanted chikungunya virus-like particle vaccine: A randomised, double-blind, parallel-group, phase 2 trial. Lancet Infect. Dis. 2022, 22, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Ramsauer, K.; Schwameis, M.; Firbas, C.; Mullner, M.; Putnak, R.J.; Thomas, S.J.; Despres, P.; Tauber, E.; Jilma, B.; Tangy, F. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: A randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect. Dis. 2015, 15, 519–527. [Google Scholar] [CrossRef]
- Reisinger, E.C.; Tschismarov, R.; Beubler, E.; Wiedermann, U.; Firbas, C.; Loebermann, M.; Pfeiffer, A.; Muellner, M.; Tauber, E.; Ramsauer, K. Immunogenicity, safety, and tolerability of the measles-vectored chikungunya virus vaccine MV-CHIK: A double-blind, randomised, placebo-controlled and active-controlled phase 2 trial. Lancet 2019, 392, 2718–2727. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.H.; Teo, T.H.; Utt, A.; Tan, J.J.; Amrun, S.N.; Abu Bakar, F.; Yee, W.X.; Becht, E.; Lee, C.Y.; Lee, B.; et al. Mutating chikungunya virus non-structural protein produces potent live-attenuated vaccine candidate. EMBO Mol. Med. 2019, 11, e10092. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Liu, X.; Zaid, A.; Goh, L.Y.; Hobson-Peters, J.; Hall, R.A.; Merits, A.; Mahalingam, S. Mutation of the N-terminal region of chikungunya virus capsid protein: Implications for vaccine design. mBio 2017, 8, e01970-16. [Google Scholar] [CrossRef] [Green Version]
- Hallengard, D.; Kakoulidou, M.; Lulla, A.; Kummerer, B.M.; Johansson, D.X.; Mutso, M.; Lulla, V.; Fazakerley, J.K.; Roques, P.; Le Grand, R.; et al. Novel attenuated chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J. Virol. 2014, 88, 2858–2866. [Google Scholar] [CrossRef] [Green Version]
- Abeyratne, E.; Freitas, J.R.; Zaid, A.; Mahalingam, S.; Taylor, A. Attenuation and stability of CHIKV-NoLS, a live-attenuated chikungunya virus vaccine candidate. Vaccines 2018, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Roques, P.; Ljungberg, K.; Kummerer, B.M.; Gosse, L.; Dereuddre-Bosquet, N.; Tchitchek, N.; Hallengard, D.; Garcia-Arriaza, J.; Meinke, A.; Esteban, M.; et al. Attenuated and vectored vaccines protect nonhuman primates against chikungunya virus. JCI Insight 2017, 2, e83527. [Google Scholar] [CrossRef]
- Wressnigg, N.; Hochreiter, R.; Zoihsl, O.; Fritzer, A.; Bezay, N.; Klingler, A.; Lingnau, K.; Schneider, M.; Lundberg, U.; Meinke, A.; et al. Single-shot live-attenuated chikungunya vaccine in healthy adults: A phase 1, randomised controlled trial. Lancet Infect. Dis. 2020, 20, 1193–1203. [Google Scholar] [CrossRef]
- Valneva Successfully Completes Pivotal Phase 3 Trial of Single-Shot Chikungunya Vaccine Candidate. Available online: https://valneva.com/press-release/valneva-successfully-completes-pivotal-phase-3-trial-of-single-shot-chikungunya-vaccine-candidate/ (accessed on 14 December 2022).
- Roques, P.; Fritzer, A.; Dereuddre-Bosquet, N.; Wressnigg, N.; Hochreiter, R.; Bossevot, L.; Pascal, Q.; Guehenneux, F.; Bitzer, A.; Corbic Ramljak, I.; et al. Effectiveness of CHIKV vaccine VLA1553 demonstrated by passive transfer of human sera. JCI Insight 2022, 7, e160173. [Google Scholar] [CrossRef] [PubMed]
- Edelman, R.; Tacket, C.O.; Wasserman, S.S.; Bodison, S.A.; Perry, J.G.; Mangiafico, J.A. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am. J. Trop. Med. Hyg. 2000, 62, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.M.; Liu, H.; Riemersma, K.K.; Ball, E.E.; Coffey, L.L. Engineering a fidelity-variant live-attenuated vaccine for chikungunya virus. NPJ Vaccines 2020, 5, 97. [Google Scholar] [CrossRef]
- Lentscher, A.J.; McAllister, N.; Griswold, K.A.; Martin, J.L.; Welsh, O.L.; Sutherland, D.M.; Silva, L.A.; Dermody, T.S. Chikungunya virus vaccine candidate incorporating synergistic mutations is attenuated and protects against virulent virus challenge. J. Infect. Dis. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Voigt, E.A.; Fuerte-Stone, J.; Granger, B.; Archer, J.; Van Hoeven, N. Live-attenuated RNA hybrid vaccine technology provides single-dose protection against chikungunya virus. Mol. Ther. 2021, 29, 2782–2793. [Google Scholar] [CrossRef]
- Abeyratne, E.; Tharmarajah, K.; Freitas, J.R.; Mostafavi, H.; Mahalingam, S.; Zaid, A.; Zaman, M.; Taylor, A. Liposomal delivery of the RNA genome of a live-attenuated chikungunya virus vaccine candidate provides local, but not systemic protection after one dose. Front. Immunol. 2020, 11, 304. [Google Scholar] [CrossRef]
- Rayner, J.O.; Kim, J.H.; Roberts, R.W.; Wood, R.R.; Fouty, B.; Solodushko, V. Evaluation of DNA-launched virus-like particle vaccines in an immune competent mouse model of chikungunya virus infection. Vaccines 2021, 9, 345. [Google Scholar] [CrossRef]
- Akahata, W.; Yang, Z.Y.; Andersen, H.; Sun, S.; Holdaway, H.A.; Kong, W.P.; Lewis, M.G.; Higgs, S.; Rossmann, M.G.; Rao, S.; et al. A virus-like particle vaccine for epidemic chikungunya virus protects nonhuman primates against infection. Nat. Med. 2010, 16, 334–338. [Google Scholar] [CrossRef] [Green Version]
- Goo, L.; Dowd, K.A.; Lin, T.Y.; Mascola, J.R.; Graham, B.S.; Ledgerwood, J.E.; Pierson, T.C. A virus-like particle vaccine elicits broad neutralizing antibody responses in humans to all chikungunya virus genotypes. J. Infect. Dis. 2016, 214, 1487–1491. [Google Scholar] [CrossRef] [Green Version]
- Lundstrom, K. Viral vectors for COVID-19 vaccine development. Viruses 2021, 13, 317. [Google Scholar] [CrossRef] [PubMed]
- Brandler, S.; Ruffie, C.; Combredet, C.; Brault, J.B.; Najburg, V.; Prevost, M.C.; Habel, A.; Tauber, E.; Despres, P.; Tangy, F. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine 2013, 31, 3718–3725. [Google Scholar] [CrossRef]
- Rossi, S.L.; Comer, J.E.; Wang, E.; Azar, S.R.; Lawrence, W.S.; Plante, J.A.; Ramsauer, K.; Schrauf, S.; Weaver, S.C. Immunogenicity and efficacy of a measles virus-vectored chikungunya vaccine in nonhuman primates. J. Infect. Dis. 2019, 220, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Camacho, C.; Kim, Y.C.; Blight, J.; Lazaro Moreli, M.; Montoya-Diaz, E.; Huiskonen, J.T.; Kummerer, B.M.; Reyes-Sandoval, A. Assessment of immunogenicity and neutralisation efficacy of viral-vectored vaccines against chikungunya virus. Viruses 2019, 11, 322. [Google Scholar] [CrossRef] [Green Version]
- Dora, E.G.; Rossi, S.L.; Weaver, S.C.; Tucker, S.N.; Mateo, R. An adjuvanted adenovirus 5-based vaccine elicits neutralizing antibodies and protects mice against chikungunya virus-induced footpad swelling. Vaccine 2019, 37, 3146–3150. [Google Scholar] [CrossRef] [PubMed]

| Signs and Symptoms | Proportion | Reference |
|---|---|---|
| Fever | 91% | [132] |
| Myalgia | 64.9% | [133] |
| 61.0% | [132] | |
| Arthralgia | 100% | [133] |
| 86% | [132] | |
| Arthralgia and myalgia | 82% | [134] |
| Arthritis | 58% | [134] |
| 56% | [132] | |
| Back pain | 55.0% | [132] |
| Rash | 54% | [134] |
| 70.05% | [133] | |
| Pruritus | 61.9% | [133] |
| 12% | [132] | |
| Conjunctivitis | 4.2% | [136] |
| 21% | [132] | |
| Non-severe hemorrhage | 5% | [135] |
| Headache | 47.1% | [135] |
| 74% | [134] | |
| 69% | [133] | |
| Fatigue | 66% | [134] |
| Nausea | 62% | [134] |
| 47% | [132] | |
| Vomiting | 60% | [134] |
| 21.0% | [132] | |
| Diarrhea | 12.0% | [132] |
| Retroorbital pain | 18.0% | [132] |
| Abdominal pain | 19.0% | [132] |
| Photophobia | 9.0% | [132] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hakim, M.S.; Aman, A.T. Understanding the Biology and Immune Pathogenesis of Chikungunya Virus Infection for Diagnostic and Vaccine Development. Viruses 2023, 15, 48. https://doi.org/10.3390/v15010048
Hakim MS, Aman AT. Understanding the Biology and Immune Pathogenesis of Chikungunya Virus Infection for Diagnostic and Vaccine Development. Viruses. 2023; 15(1):48. https://doi.org/10.3390/v15010048
Chicago/Turabian StyleHakim, Mohamad S., and Abu T. Aman. 2023. "Understanding the Biology and Immune Pathogenesis of Chikungunya Virus Infection for Diagnostic and Vaccine Development" Viruses 15, no. 1: 48. https://doi.org/10.3390/v15010048
APA StyleHakim, M. S., & Aman, A. T. (2023). Understanding the Biology and Immune Pathogenesis of Chikungunya Virus Infection for Diagnostic and Vaccine Development. Viruses, 15(1), 48. https://doi.org/10.3390/v15010048





