The Hypersensitive Response to Plant Viruses
Abstract
:1. Introduction
2. Resistance Genes to Viral Infection
2.1. Involvement of LRR-RLK Encoding Genes in Response to Viral Infection
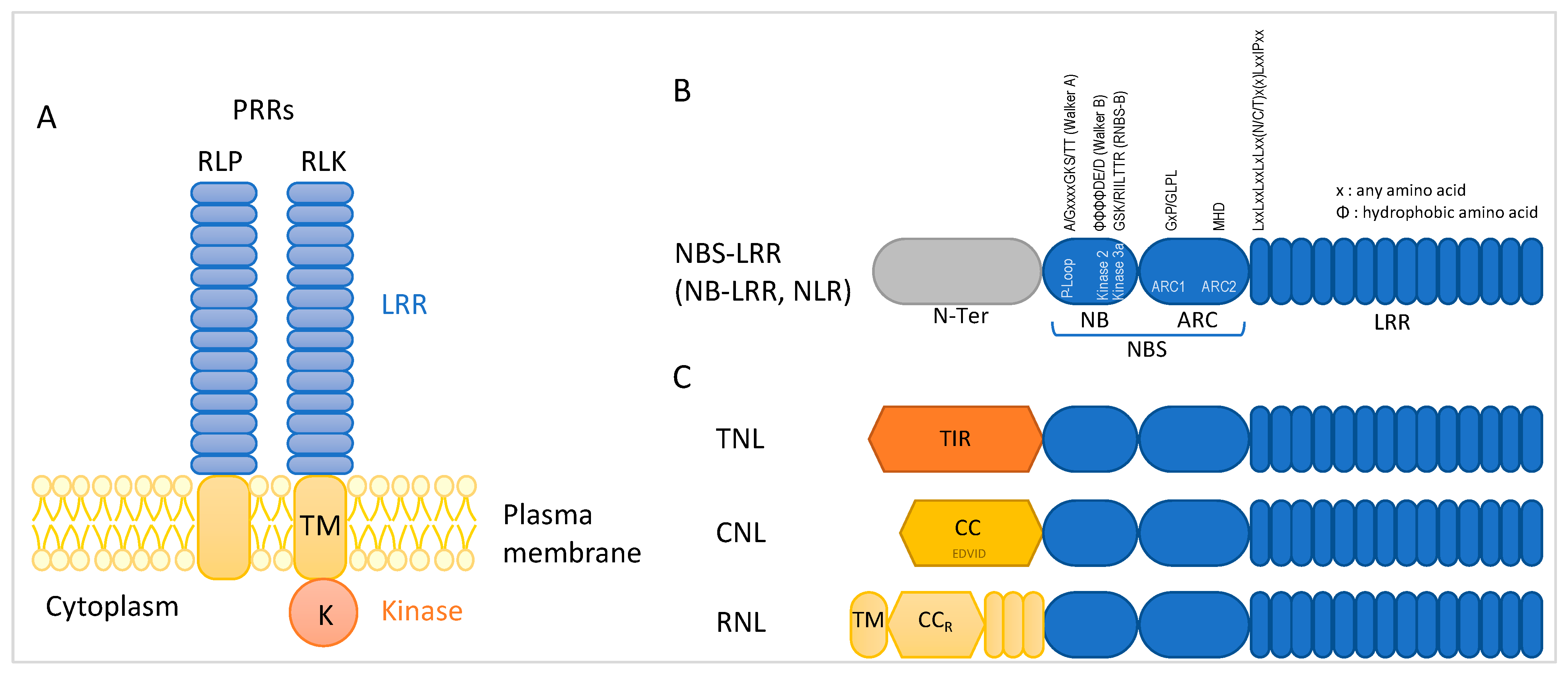
2.2. NBS-LRR Encoding R Genes
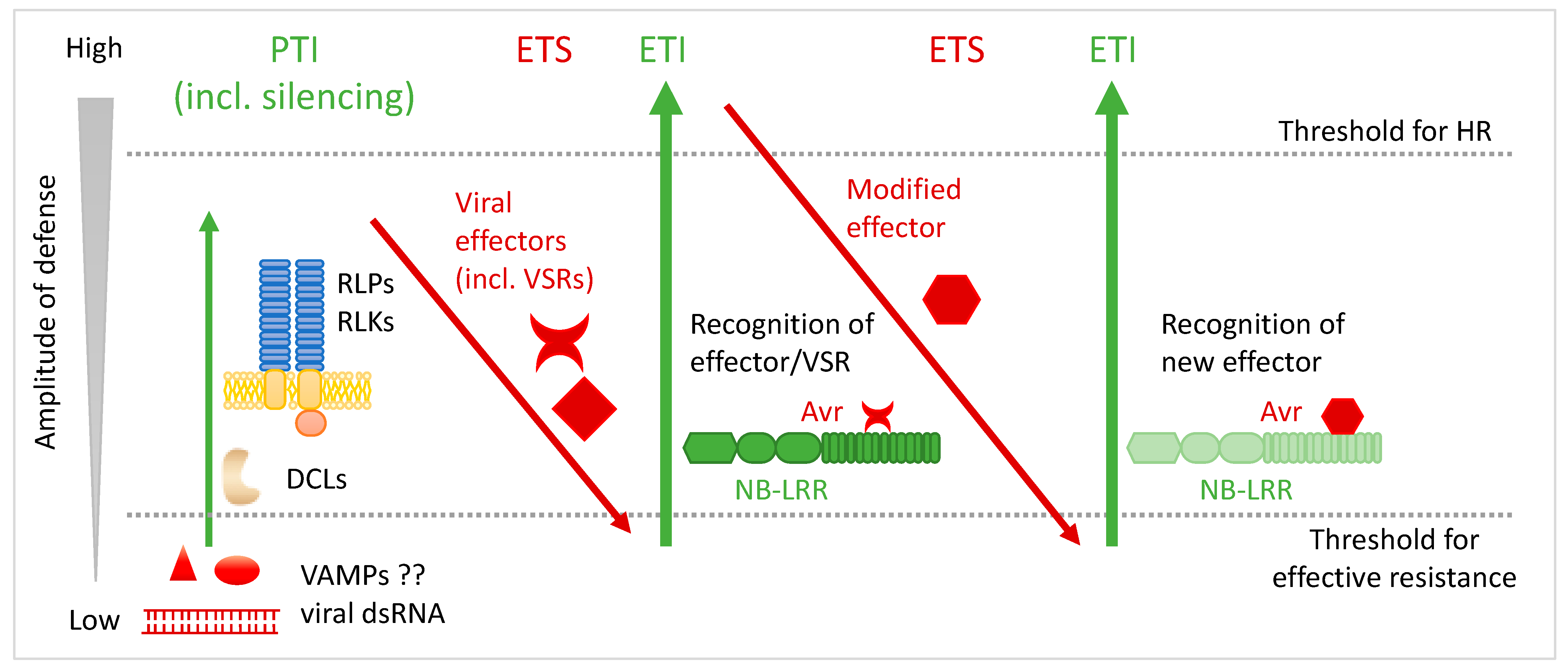
3. Avr Factors and Avr–R Recognition Models
4. Important Advances in the Comprehension of HR
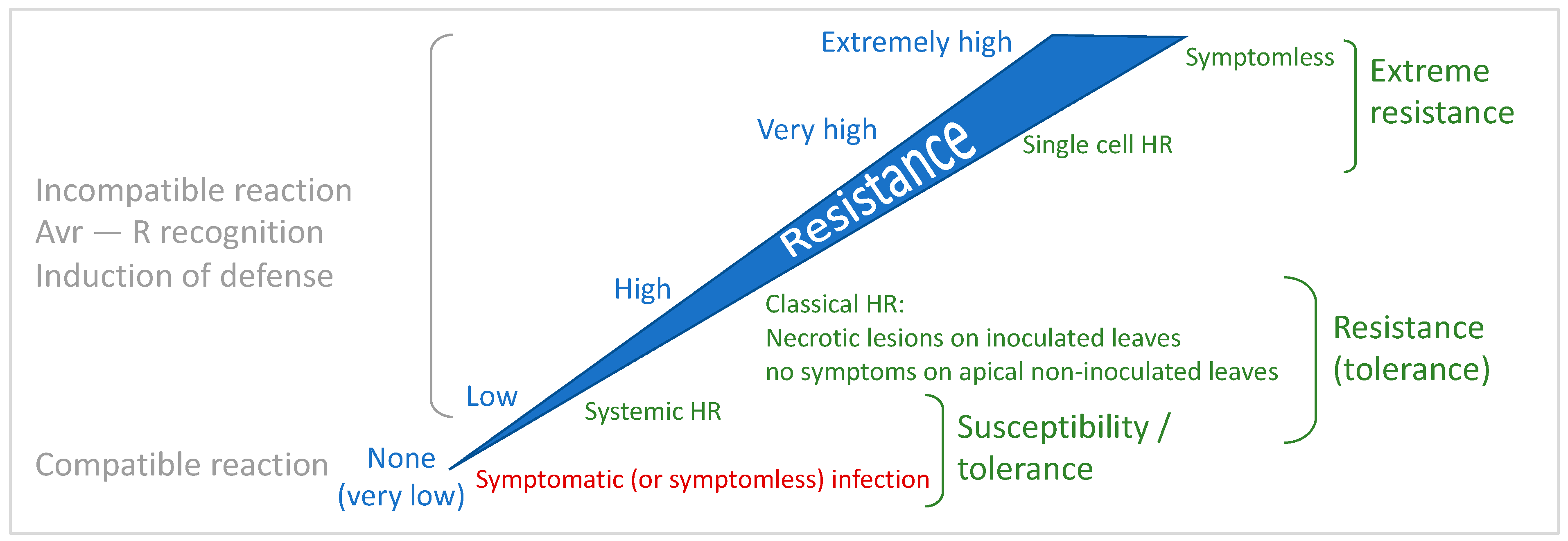
4.1. The Continuum of Resistance: Uncoupling Cell Death and Resistance
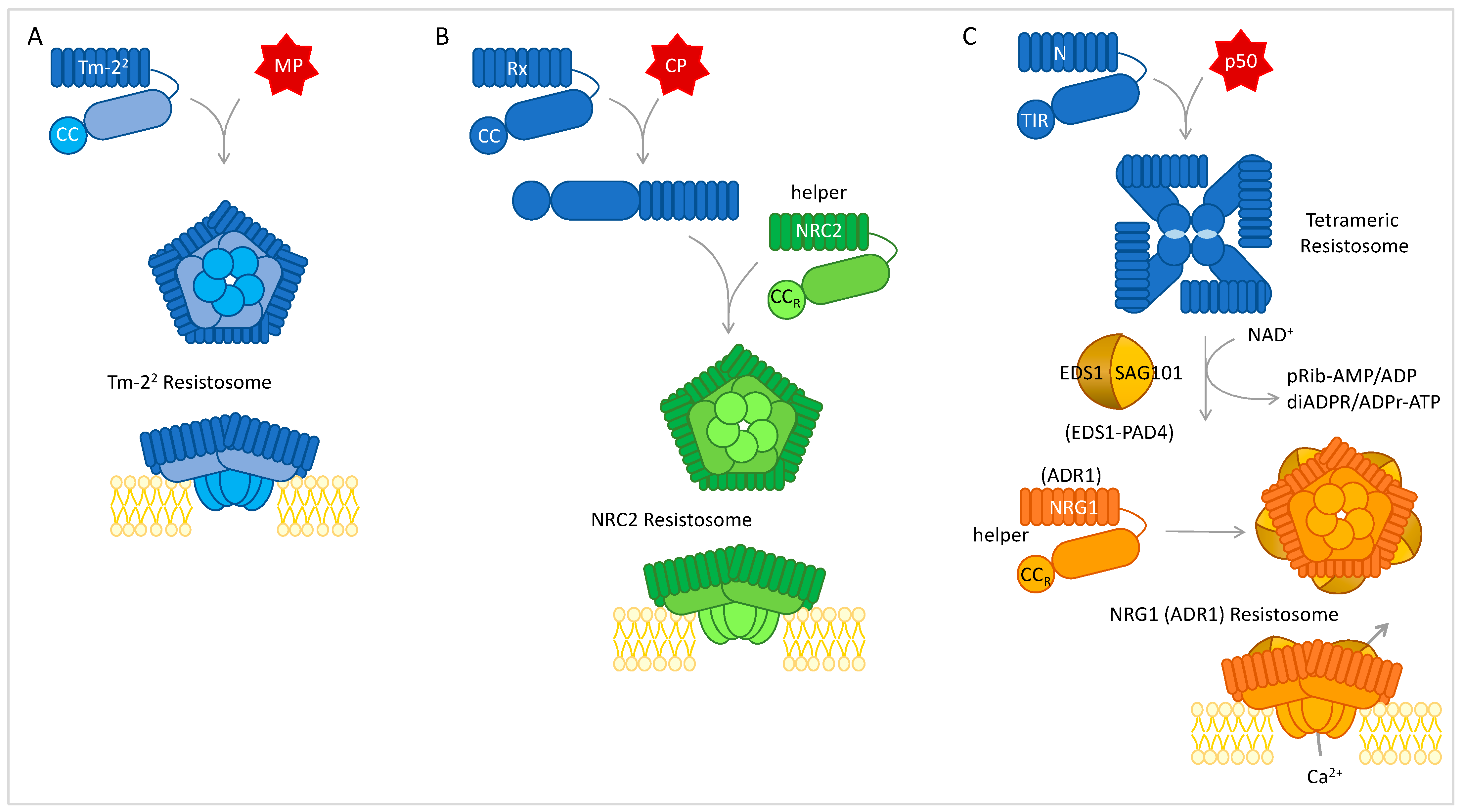
4.2. Resistosomes, NLR Networks, and Convergence
4.3. Subcellular Localization
4.4. HR Is Controlled by UPS and Autophagy
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hilaire, J.; Tindale, S.; Jones, G.; Pingarron-Cardenas, G.; Bačnik, K.; Ojo, M.; Frewer, L.J. Risk perception associated with an emerging agri-food risk in Europe: Plant viruses in agriculture. Agric. Food Secur. 2022, 11, 21. [Google Scholar] [CrossRef]
- Jones, R.A.C. Global plant virus disease pandemics and epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Naidu, R.A. Global dimensions of plant virus diseases: Current status and future perspectives. Annu. Rev. Virol. 2019, 6, 387–409. [Google Scholar] [CrossRef]
- Fuchs, M. Pyramiding resistance-conferring gene sequences in crops. Curr. Opin. Virol. 2017, 26, 36–42. [Google Scholar] [CrossRef]
- Schmitt-Keichinger, C. Manipulating cellular factors to combat viruses: A case study from the plant eukaryotic translation initiation factors eIF4. Front. Microbiol. 2019, 10, 17. [Google Scholar] [CrossRef]
- Sanfaçon, H. Plant translation factors and virus resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef]
- Ding, S.-W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Lopez-Gomollon, S.; Baulcombe, D.C. Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 2022, 23, 645–662. [Google Scholar] [CrossRef]
- Zhu, H.; Guo, H. The role of virus-derived small interfering RNAs in RNA silencing in plants. Sci. China Life Sci. 2012, 55, 119–125. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Ravelonandro, M.; Scorza, R.; Callahan, A.; Levy, L.; Jacquet, C.; Monsion, M.; Damsteegt, V. The use of transgenic fruit trees as a resistance strategy for virus epidemics: The plum pox (sharka) model. Virus Res. 2000, 71, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Gonsalves, D. Safety of virus-resistant transgenic plants two decades after their introduction: Lessons from realistic field risk assessment studies. Annu. Rev. Phytopathol. 2007, 45, 173–202. [Google Scholar] [CrossRef]
- Singh, K.; Dardick, C.; Kumar Kundu, J. RNAi-mediated resistance against viruses in perennial fruit plants. Plants 2019, 8, 359. [Google Scholar] [CrossRef] [PubMed]
- Runo, S.; Alakonya, A.; Machuka, J.; Sinha, N. RNA interference as a resistance mechanism against crop parasites in Africa: A ‘Trojan horse’ approach. Pest Manag. Sci. 2011, 67, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, Z.; Li, Y.; Wu, J. Biogenesis, function, and applications of virus-derived small RNAs in plants. Front. Microbiol. 2015, 6, 1237. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Trivedi, P.K. Artificial microRNA mediated gene silencing in plants: Progress and perspectives. Plant Mol. Biol. 2014, 86, 1–18. [Google Scholar] [CrossRef]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNAs for plant virus resistance. In Antiviral Resistance in Plants; Watson, J.M., Wang, M.-B., Eds.; Humana Press: Totowa, NJ, USA, 2012; Volume 894, pp. 209–222. [Google Scholar]
- Kumar, K.K.; Varanavasiappan, S.; Arul, L.; Kokiladevi, E.; Sudhakar, D. Strategies for efficient RNAi-based gene silencing of viral genes for disease resistance in plants. In Plant Gene Silencing: Methods and Protocols; Mysore, K.S., Senthil-Kumar, M., Eds.; Springer: New York, NY, USA, 2022; pp. 23–35. [Google Scholar]
- Teixeira, R.M.; Ferreira, M.A.; Raimundo, G.A.S.; Loriato, V.A.P.; Reis, P.A.B.; Fontes, E.P.B. Virus perception at the cell surface: Revisiting the roles of receptor-like kinases as viral pattern recognition receptors. Mol. Plant Pathol. 2019, 20, 1196–1202. [Google Scholar] [CrossRef]
- Moffett, P. Mechanisms of recognition in dominant R gene mediated resistance. In Advances in Virus Research; Moffett, P., Gad, L., John, P.C., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 75, pp. 1–33. [Google Scholar]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef]
- Zvereva, A.S.; Pooggin, M.M. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Fátyol, K.; Fekete, K.A.; Ludman, M. Double-stranded-RNA-binding protein 2 participates in antiviral defense. J. Virol. 2020, 94, e00017-20. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef]
- Kørner, C.J.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 2013, 26, 1271–1280. [Google Scholar] [CrossRef]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef]
- Nicaise, V.; Candresse, T. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol. Plant Pathol. 2017, 18, 878–886. [Google Scholar] [CrossRef]
- Künstler, A.; Bacsó, R.; Gullner, G.; Hafez, Y.M.; Király, L. Staying alive—Is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 2016, 93, 75–84. [Google Scholar] [CrossRef]
- Niehl, A.; Heinlein, M. Perception of double-stranded RNA in plant antiviral immunity. Mol. Plant Pathol. 2019, 20, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral protein suppresses oxidative burst and salicylic acid-dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.; Teixeira, R.M.; Fontes, E.P.B. Geminivirus-host interactions: Action and reaction in receptor-mediated antiviral immunity. Viruses 2021, 13, 840. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Wei, M.; Li, G.; Lei, R.; Qiu, Y.; Wang, C.; Li, Z.H.; Zhu, S. The cucumber mosaic virus movement protein suppresses PAMP-triggered immune responses in Arabidopsis and tobacco. Biochem. Biophys. Res. Commun. 2018, 498, 395–401. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, D.; Wang, X.; Zhang, X.; Wen, Z.; Zhang, Q.; Li, D.; Dinesh-Kumar, S.P.; Zhang, Y. Coat proteins of necroviruses target 14-3-3a to subvert MAPKKKalpha-mediated antiviral immunity in plants. Nat. Commun. 2022, 13, 716. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef]
- Lu, Y.; Tsuda, K. Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol. Plant Microbe Interact. 2021, 34, 3–14. [Google Scholar] [CrossRef]
- Lou, L.; Su, X.; Liu, X.; Liu, Z. Transcriptome analysis of Luffa cylindrica (L.) Roem response to infection with cucumber mosaic virus (CMV). Gene 2020, 737, 144451. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, T.; Jia, Z.; Jia, X.; Liu, Y.; Xuan, J.; Wang, G.; Zhang, F. Transcriptome analysis reveals a comprehensive virus resistance response mechanism in pecan infected by a novel badnavirus pecan virus. Int. J. Mol. Sci. 2022, 23, 13576. [Google Scholar] [CrossRef]
- Khalilzadeh, M.; Weber, K.C.; Dutt, M.; El-Mohtar, C.A.; Levy, A. Comparative transcriptome analysis of Citrus macrophylla tree infected with citrus tristeza virus stem pitting mutants provides new insight into the role of phloem regeneration in stem pitting disease. Front. Plant Sci. 2022, 13, 987831. [Google Scholar] [CrossRef]
- Khan, A.; Johnson George, K.; Jasrotia, R.S.; Aravind, S.; Angadi, U.B.; Iquebal, M.A.; Manju, K.P.; Jaiswal, S.; Umadevi, P.; Rai, A.; et al. Plant virus interaction mechanism and associated pathways in mosaic disease of small cardamom (Elettaria cardamomum Maton) by RNA-Seq approach. Genomics 2020, 112, 2041–2051. [Google Scholar] [CrossRef] [PubMed]
- Das, P.P.; Macharia, M.W.; Lin, Q.; Wong, S.M. In planta proximity-dependent biotin identification (BioID) identifies a TMV replication co-chaperone NbSGT1 in the vicinity of 126 kDa replicase. J. Proteom. 2019, 204, 103402. [Google Scholar] [CrossRef] [PubMed]
- Ariza-Suarez, D.; Keller, B.; Spescha, A.; Aparicio, J.S.; Mayor, V.; Portilla-Benavides, A.E.; Buendia, H.F.; Bueno, J.M.; Studer, B.; Raatz, B. Genetic analysis of resistance to bean leaf crumple virus identifies a candidate LRR-RLK gene. Plant J. 2023, 114, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Boualem, A.; Dogimont, C.; Bendahmane, A. The battle for survival between viruses and their host plants. Curr. Opin. Virol. 2016, 17, 32–38. [Google Scholar] [CrossRef]
- Prasad, L.; Katoch, S.; Shahid, S. Microbial interaction mediated programmed cell death in plants. 3 Biotech 2022, 12, 43. [Google Scholar] [CrossRef]
- Dinesh-Kumar, S.P.; Whitham, S.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. Transposon tagging of tobacco mosaic virus resistance gene N: Its possible role in the TMV-N-mediated signal transduction pathway. Proc. Natl. Acad. Sci. USA 1995, 92, 4175–4180. [Google Scholar] [CrossRef]
- van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef]
- Martin, E.C.; Spiridon, L.; Goverse, A.; Petrescu, A.J. NLRexpress-A bundle of machine learning motif predictors reveals motif stability underlying plant Nod-like receptors diversity. Front. Plant Sci. 2022, 13, 975888. [Google Scholar] [CrossRef]
- Shao, Z.-Q.; Xue, J.-Y.; Wu, P.; Zhang, Y.-M.; Wu, Y.; Hang, Y.-Y.; Wang, B.; Chen, J.-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef]
- Collier, S.M.; Hamel, L.-P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Barragan, C.A.; Wu, R.; Kim, S.T.; Xi, W.; Habring, A.; Hagmann, J.; Van de Weyer, A.L.; Zaidem, M.; Ho, W.W.H.; Wang, G.; et al. RPW8/HR repeats control NLR activation in Arabidopsis thaliana. PLoS Genet. 2019, 15, e1008313. [Google Scholar] [CrossRef]
- Wu, C.H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Barragan, A.C.; Weigel, D. Plant NLR diversity: The known unknowns of pan-NLRomes. Plant Cell 2021, 33, 814–831. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, C.E.; Astua-Monge, G.; Jones, V.; Plyler, T.R.; Sakiyama, N.S.; Mackenzie, S.A. Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 2006, 172, 1229–1242. [Google Scholar] [CrossRef]
- Baebler, Š.; Coll, A.; Gruden, K. Plant molecular responses to potato virus Y: A continuum of outcomes from sensitivity and tolerance to resistance. Viruses 2020, 12, 217. [Google Scholar] [CrossRef]
- Torrance, L.; Cowan, G.H.; McLean, K.; MacFarlane, S.; Al-Abedy, A.N.; Armstrong, M.; Lim, T.Y.; Hein, I.; Bryan, G.J. Natural resistance to potato virus Y in Solanum tuberosum group Phureja. Theor. Appl. Genet. 2020, 133, 967–980. [Google Scholar] [CrossRef]
- Ellis, M.H.; Stiller, W.N.; Phongkham, T.; Tate, W.A.; Gillespie, V.J.; Gapare, W.J.; Zhu, Q.-H.; Llewellyn, D.J.; Wilson, I.W. Molecular mapping of bunchy top disease resistance in Gossypium hirsutum L. Euphytica 2016, 210, 135–142. [Google Scholar] [CrossRef]
- Fang, D.D.; Xiao, J.; Canci, P.C.; Cantrell, R.G. A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2010, 120, 943–953. [Google Scholar] [CrossRef]
- Lu, X.; Li, Z.; Huang, W.; Wang, S.; Zhang, S.; Li, F.; Zhang, H.; Sun, R.; Li, G.; Zhang, S. Mapping and identification of a new potential dominant resistance gene to turnip mosaic virus in Brassica rapa. Planta 2022, 256, 66. [Google Scholar] [CrossRef]
- Jupe, F.; Witek, K.; Verweij, W.; Sliwka, J.; Pritchard, L.; Etherington, G.J.; Maclean, D.; Cock, P.J.; Leggett, R.M.; Bryan, G.J.; et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 2013, 76, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Witek, K.; Jupe, F.; Witek, A.I.; Baker, D.; Clark, M.D.; Jones, J.D. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 2016, 34, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.G.; Hennig, J. Extreme resistance to potato virus Y in potato carrying the Ry(sto) gene is mediated by a TIR-NLR immune receptor. Plant Biotechnol. J. 2020, 18, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.G.; de Haan, P.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1039–1051. [Google Scholar] [CrossRef]
- Brendolise, C.; Montefiori, M.; Dinis, R.; Peeters, N.; Storey, R.D.; Rikkerink, E.H. A novel hairpin library-based approach to identify NBS-LRR genes required for effector-triggered hypersensitive response in Nicotiana benthamiana. Plant Methods 2017, 13, 32. [Google Scholar] [CrossRef]
- Li, N.; Yin, J.L.; Li, C.; Wang, D.G.; Yang, Y.Q.; Karthikeyan, A.; Luan, H.X.; Zhi, H.J. NB-LRR gene family required for Rsc4-mediated resistance to soybean mosaic virus. Crop Pasture Sci. 2016, 67, 541–552. [Google Scholar] [CrossRef]
- Yin, J.; Wang, L.; Jin, T.; Nie, Y.; Liu, H.; Qiu, Y.; Yang, Y.; Li, B.; Zhang, J.; Wang, D.; et al. A cell wall-localized NLR confers resistance to soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol. Plant 2021, 14, 1881–1900. [Google Scholar] [CrossRef]
- Bendahmane, A.; Querci, M.; Kanyuka, K.; Baulcombe, D.C. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 2000, 21, 73–81. [Google Scholar] [CrossRef]
- Tran, P.-T.; Choi, H.; Kim, S.-B.; Lee, H.-A.; Choi, D.; Kim, K.-H. A simple method for screening of plant NBS-LRR genes that confer a hypersensitive response to plant viruses and its application for screening candidate pepper genes against Pepper mottle virus. J. Virol. Methods 2014, 201, 57–64. [Google Scholar] [CrossRef]
- Tran, P.-T.; Choi, H.; Choi, D.; Kim, K.-H. Molecular characterization of Pvr9 that confers a hypersensitive response to pepper mottle virus (a potyvirus) in Nicotiana benthamiana. Virology 2015, 481, 113–123. [Google Scholar] [CrossRef]
- Tomita, R.; Sekine, K.-T.; Mizumoto, H.; Sakamoto, M.; Murai, J.; Kiba, A.; Hikichi, Y.; Suzuki, K.; Kobayashi, K. Genetic basis for the hierarchical interaction between tobamovirus spp. and L resistance gene alleles from different pepper species. Mol. Plant Microbe Interact. 2011, 24, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.T.; Tomita, R.; Takeuchi, S.; Atsumi, G.; Saitoh, H.; Mizumoto, H.; Kiba, A.; Yamaoka, N.; Nishiguchi, M.; Hikichi, Y.; et al. Functional differentiation in the leucine-rich repeat domains of closely related plant virus-resistance proteins that recognize common Avr proteins. Mol. Plant Microbe Interact. 2012, 25, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-B.; Kang, W.-H.; Huy, H.N.; Yeom, S.-I.; An, J.-T.; Kim, S.; Kang, M.-Y.; Kim, H.J.; Jo, Y.D.; Ha, Y.; et al. Divergent evolution of multiple virus-resistance genes from a progenitor in Capsicum spp. New Phytol. 2017, 213, 886–899. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef]
- Padgett, H.S.; Watanabe, Y.; Beachy, R.N. Identification of the TMV replicase sequence that activates the N gene-mediated hypersensitive response. Mol. Plant Microbe Interact. 1997, 10, 709–715. [Google Scholar] [CrossRef]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef]
- Bendahmane, A.; Köhm, B.A.; Dedi, C.; Baulcombe, D.C. The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 1995, 8, 933–941. [Google Scholar] [CrossRef]
- Querci, M.; Baulcombe, D.C.; Goldbach, R.W.; Salazar, L.F. Analysis of the resistance-breaking determinants of potato virus X (PVX) strain HB on different potato genotypes expressing extreme resistance to PVX. Phytopathlogy 1995, 85, 1003–1010. [Google Scholar] [CrossRef]
- Cooley, M.B.; Pathirana, S.; Wu, H.J.; Kachroo, P.; Klessig, D.F. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 2000, 12, 663–676. [Google Scholar] [CrossRef]
- Spassova, M.I.; Prins, T.W.; Folkertsma, R.T.; Klein-Lankhorst, R.M.; Hille, J.; Goldbach, R.W.; Prins, M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 2001, 7, 151–161. [Google Scholar] [CrossRef]
- Peiró, A.; Cañizares, M.C.; Rubio, L.; López, C.; Moriones, E.; Aramburu, J.; Sánchez-Navarro, J. The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol. Plant Pathol. 2014, 15, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Miller, J.; Nozaki, Y.; Takeda, M.; Shah, J.; Hase, S.; Ikegami, M.; Ehara, Y.; Dinesh-Kumar, S.P. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 2002, 32, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Suzuki, M.; Natsuaki, K.; Shigyo, T.; Hino, K.; Teraoka, T.; Hosokawa, D.; Ehara, Y. Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus-Infected Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 340–347. [Google Scholar] [CrossRef]
- Vidal, S.; Cabrera, H.; Andersson, R.A.; Fredriksson, A.; Valkonen, J.P. Potato gene Y-1 is an N gene homolog that confers cell death upon infection with potato virus Y. Mol. Plant Microbe Interact. 2002, 15, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Pfitzner, A.J. Tm-2(2) resistance in tomato requires recognition of the carboxy terminus of the movement protein of tomato mosaic virus. Mol. Plant Microbe Interact. 1998, 11, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-S.; Rojas, M.R.; Lee, J.-Y.; Lee, S.-W.; Jeon, J.-S.; Ronald, P.; Lucas, W.J.; Gilbertson, R.L. A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc. Natl. Acad. Sci. USA 2006, 103, 11856–11861. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-S.; Jeon, J.-S.; Rojas, M.R.; Gilbertson, R.L. Characterization of a novel Toll/interleukin-1 receptor (TIR)-TIR gene differentially expressed in common bean (Phaseolus vulgaris cv. Othello) undergoing a defence response to the geminivirus Bean dwarf mosaic virus. Mol. Plant Pathol. 2007, 8, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramirez, E.R.; Sudarshana, M.R.; Lucas, W.J.; Gilbertson, R.L. Bean dwarf mosaic virus BV1 protein is a determinant of the hypersensitive response and avirulence in Phaseolus vulgaris. Mol. Plant Microbe Interact. 2000, 13, 1184–1194. [Google Scholar] [CrossRef]
- Ma, J.; Hou, X.; Xiao, D.; Qi, L.; Wang, F.; Sun, F.; Wang, Q. Cloning and characterization of the BcTuR3 gene related to resistance to turnip mosaic virus (TuMV) from non-heading Chinese cabbage. Plant Mol. Biol. Rep. 2010, 28, 588–596. [Google Scholar] [CrossRef]
- Berzal-Herranz, A.; de la Cruz, A.; Tenllado, F.; Díaz-Ruíz, J.R.; López, L.; Sanz, A.I.; Vaquero, C.; Serra, M.T.; García-Luque, I. The Capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology 1995, 209, 498–505. [Google Scholar] [CrossRef]
- Culver, J.N.; Dawson, W.O. Tobacco mosaic virus coat protein: An elicitor of the hypersensitive reaction but not required for the development of mosaic symptoms in Nicotiana sylvestris. Virology 1989, 173, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Maiti, S.; Paul, S.; Pal, A. Isolation, characterization, and structure analysis of a non-TIR-NBS-LRR encoding candidate gene from MYMIV-resistant Vigna mungo. Mol. Biotechnol. 2012, 52, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Brotman, Y.; Normantovich, M.; Goldenberg, Z.; Zvirin, Z.; Kovalski, I.; Stovbun, N.; Doniger, T.; Bolger, A.M.; Troadec, C.; Bendahmane, A.; et al. Dual resistance of melon to Fusarium oxysporum races 0 and 2 and to papaya ring-spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol. Plant 2013, 6, 235–238. [Google Scholar] [CrossRef]
- Nizan, S.; Amitzur, A.; Dahan-Meir, T.; Benichou, J.I.C.; Bar-Ziv, A.; Perl-Treves, R. Mutagenesis of the melon Prv gene by CRISPR/Cas9 breaks papaya ringspot virus resistance and generates an autoimmune allele with constitutive defense responses. J. Exp. Bot. 2023, 74, 4579–4596. [Google Scholar] [CrossRef]
- Jin, M.; Lee, S.S.; Ke, L.; Kim, J.S.; Seo, M.S.; Sohn, S.H.; Park, B.S.; Bonnema, G. Identification and mapping of a novel dominant resistance gene, TuRB07 to turnip mosaic virus in Brassica rapa. Theor. Appl. Genet. 2014, 127, 509–519. [Google Scholar] [CrossRef]
- Ma, F.F.; Wu, X.Y.; Chen, Y.X.; Liu, Y.N.; Shao, Z.Q.; Wu, P.; Wu, M.; Liu, C.C.; Wu, W.P.; Yang, J.Y.; et al. Fine mapping of the Rsv1-h gene in the soybean cultivar Suweon 97 that confers resistance to two Chinese strains of the soybean mosaic virus. Theor. Appl. Genet. 2016, 129, 2227–2236. [Google Scholar] [CrossRef] [PubMed]
- Eggenberger, A.L.; Hajimorad, M.R.; Hill, J.H. Gain of virulence on Rsv1-genotype soybean by an avirulent soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; An, J.; Kang, W.H.; Jahn, M.; Kang, B.C. Fine mapping of the dominant potyvirus resistance gene Pvr7 reveals a relationship with Pvr4 in Capsicum annuum. Phytopathology 2018, 108, 142–148. [Google Scholar] [CrossRef]
- Kim, S.B.; Lee, H.Y.; Seo, S.; Lee, J.H.; Choi, D. RNA-dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS ONE 2015, 10, e0119639. [Google Scholar] [CrossRef]
- de Ronde, D.; Butterbach, P.; Lohuis, D.; Hedil, M.; van Lent, J.W.; Kormelink, R. Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the tomato spotted wilt virus. Mol. Plant Pathol. 2013, 14, 405–415. [Google Scholar] [CrossRef]
- Capistrano-Gossmann, G.G.; Ries, D.; Holtgrawe, D.; Minoche, A.; Kraft, T.; Frerichmann, S.L.M.; Rosleff Soerensen, T.; Dohm, J.C.; Gonzalez, I.; Schilhabel, M.; et al. Crop wild relative populations of Beta vulgaris allow direct mapping of agronomically important genes. Nat. Commun. 2017, 8, 15708. [Google Scholar] [CrossRef]
- Wetzel, V.; Willlems, G.; Darracq, A.; Galein, Y.; Liebe, S.; Varrelmann, M. The Beta vulgaris-derived resistance gene Rz2 confers broad-spectrum resistance against soilborne sugar beet-infecting viruses from different families by recognizing triple gene block protein 1. Mol. Plant Pathol. 2021, 22, 829–842. [Google Scholar] [CrossRef]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Bai, Y.; Wolters, A.A. The NLR protein encoded by the resistance gene Ty-2 is triggered by the replication-associated protein Rep/C1 of tomato yellow leaf curl virus. Front. Plant Sci. 2020, 11, 545306. [Google Scholar] [CrossRef]
- Mestre, P.; Brigneti, G.; Baulcombe, D.C. An Ry-mediated resistance response in potato requires the intact active site of the NIa proteinase from potato virus Y. Plant J. 2000, 23, 653–661. [Google Scholar] [CrossRef]
- Sharma, N.; Sahu, P.P.; Prasad, A.; Muthamilarasan, M.; Waseem, M.; Khan, Y.; Thakur, J.K.; Chakraborty, S.; Prasad, M. The Sw5a gene confers resistance to ToLCNDV and triggers an HR response after direct AC4 effector recognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2101833118. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Shao, J.; Wang, Y.; Liu, T.; Tong, Y.; Jansky, S.; Xie, C.; Song, B.; Cai, X. Rychc confers extreme resistance to potato virus Y in potato. Cells 2022, 11, 2577. [Google Scholar] [CrossRef] [PubMed]
- Malcuit, I.; Marano, M.R.; Kavanagh, T.A.; De Jong, W.; Forsyth, A.; Baulcombe, D.C. The 25-kDa movement protein of PVX elicits Nb-mediated hypersensitive cell death in potato. Mol. Plant-Microbe Interact. 1999, 12, 536–543. [Google Scholar] [CrossRef]
- Wang, K.D.; Empleo, R.; Nguyen, T.T.; Moffett, P.; Sacco, M.A. Elicitation of hypersensitive responses in Nicotiana glutinosa by the suppressor of RNA silencing protein P0 from poleroviruses. Mol. Plant Pathol. 2015, 16, 435–448. [Google Scholar] [CrossRef]
- Agrofoglio, Y.C.; Delfosse, V.C.; Casse, M.F.; Hopp, H.E.; Bonacic Kresic, I.; Ziegler-Graff, V.; Distéfano, A.J. P0 protein of cotton leafroll dwarf virus-atypical isolate is a weak RNA silencing suppressor and the avirulence determinant that breaks the cotton Cbd gene-based resistance. Plant Pathol. 2019, 68, 1059–1071. [Google Scholar] [CrossRef]
- Palanichelvam, K.; Cole, A.B.; Shababi, M.; Schoelz, J.E. Agroinfiltration of cauliflower mosaic virus gene VI elicits hypersensitive response in Nicotiana species. Mol. Plant Microbe Interact. 2000, 13, 1275–1279. [Google Scholar] [CrossRef]
- Martin, I.R.; Vigne, E.; Berthold, F.; Komar, V.; Lemaire, O.; Fuchs, M.; Schmitt-Keichinger, C. The 50 distal amino acids of the 2AHP homing protein of Grapevine fanleaf virus elicit a hypersensitive reaction on Nicotiana occidentalis. Mol. Plant Pathol. 2018, 19, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, Y.; Yu, H.; Yuan, C.; Zeng, J.; Zhao, L.; Tong, Z.; Tao, X. Non-structural protein NSm of tomato spotted wilt virus is an avirulence factor recognized by resistance genes of tobacco and tomato via different elicitor active sites. Viruses 2018, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ren, Y.; Wang, D.; Farooq, T.; He, Z.; Zhang, C.; Li, S.; Yang, X.; Zhou, X. A group I WRKY transcription factor regulates mulberry mosaic dwarf-associated virus-triggered cell death in Nicotiana benthamiana. Mol. Plant Pathol. 2022, 23, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Hapiak, M.; Li, Y.; Agama, K.; Swade, S.; Okenka, G.; Falk, J.; Khandekar, S.; Raikhy, G.; Anderson, A.; Pollock, J.; et al. Cauliflower mosaic virus gene VI product N-terminus contains regions involved in resistance-breakage, self-association and interactions with movement protein. Virus Res. 2008, 138, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; DelGrosso, L.; Yigit, E.; Dempsey, D.A.; Klessig, D.F.; Wobbe, K.K. The amino terminus of the coat protein of turnip crinkle virus is the AVR factor recognized by resistant Arabidopsis. Mol. Plant Microbe Interact. 2000, 13, 1015–1018. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Huang, C. From player to pawn: Viral avirulence factors involved in plant immunity. Viruses 2021, 13, 688. [Google Scholar] [CrossRef]
- Sett, S.; Prasad, A.; Prasad, M. Resistance genes on the verge of plant–virus interaction. Trends Plant Sci. 2022, 27, 1242–1252. [Google Scholar] [CrossRef]
- Slootweg, E.; Koropacka, K.; Roosien, J.; Dees, R.; Overmars, H.; Lankhorst, R.K.; van Schaik, C.; Pomp, R.; Bouwman, L.; Helder, J.; et al. Sequence exchange between homologous NB-LRR genes converts virus resistance into nematode resistance, and vice versa. Plant Physiol. 2017, 175, 498–510. [Google Scholar] [CrossRef]
- Caplan, J.L.; Mamillapalli, P.; Burch-Smith, T.M.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 2008, 132, 449–462. [Google Scholar] [CrossRef]
- Li, J.; Huang, H.; Zhu, M.; Huang, S.; Zhang, W.; Dinesh-Kumar, S.P.; Tao, X. A plant immune receptor adopts a two-step recognition mechanism to enhance viral effector perception. Mol. Plant 2019, 12, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jiang, L.; Bai, B.; Zhao, W.; Chen, X.; Li, J.; Liu, Y.; Chen, Z.; Wang, B.; Wang, C.; et al. The intracellular immune receptor Sw-5b confers broad-spectrum resistance to tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- van der Hoorn, R.A.L.; Kamoun, S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef]
- Cesari, S.; Bernoux, M.; Moncuquet, P.; Kroj, T.; Dodds, P. A novel conserved mechanism for plant NLR protein pairs: The ‘integrated decoy’ hypothesis. Front. Plant Sci. 2014, 5, 606. [Google Scholar] [CrossRef] [PubMed]
- Kroj, T.; Chanclud, E.; Michel-Romiti, C.; Grand, X.; Morel, J.B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Sacco, M.A.; Mansoor, S.; Moffett, P. A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 2007, 52, 82–93. [Google Scholar] [CrossRef]
- Hao, W.; Collier, S.M.; Moffett, P.; Chai, J. Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating Protein 2 (RanGAP2). J. Biol. Chem. 2013, 288, 35868–35876. [Google Scholar] [CrossRef]
- Sun, Y.; Zhu, Y.X.; Balint-Kurti, P.J.; Wang, G.F. Fine-tuning immunity: Players and regulators for plant NLRs. Trends Plant Sci. 2020, 25, 695–713. [Google Scholar] [CrossRef]
- Baggs, E.; Dagdas, G.; Krasileva, K.V. NLR diversity, helpers and integrated domains: Making sense of the NLR IDentity. Curr. Opin. Plant Biol. 2017, 38, 59–67. [Google Scholar] [CrossRef]
- Marchal, C.; Michalopoulou, V.A.; Zou, Z.; Cevik, V.; Sarris, P.F. Show me your ID: NLR immune receptors with integrated domains in plants. Essays Biochem. 2022, 66, 527–539. [Google Scholar]
- Zhang, X.; Dodds, P.N.; Bernoux, M. What do we know about NOD-like receptors in plant immunity? Annu. Rev. Phytopathol. 2017, 55, 205–229. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Saile, S.C.; Jacob, P.; Castel, B.; Jubic, L.M.; Salas-Gonzáles, I.; Bäcker, M.; Jones, J.D.G.; Dangl, J.L.; El Kasmi, F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 2020, 18, e3000783. [Google Scholar] [CrossRef] [PubMed]
- Contreras, M.P.; Pai, H.; Tumtas, Y.; Duggan, C.; Yuen, E.L.H.; Cruces, A.V.; Kourelis, J.; Ahn, H.-K.; Lee, K.-T.; Wu, C.-H.; et al. Sensor NLR immune proteins activate oligomerization of their NRC helpers in response to plant pathogens. EMBO J. 2023, 42, e111519. [Google Scholar] [CrossRef]
- Bernoux, M.; Burdett, H.; Williams, S.J.; Zhang, X.; Chen, C.; Newell, K.; Lawrence, G.J.; Kobe, B.; Ellis, J.G.; Anderson, P.A.; et al. Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 2016, 28, 146–159. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Gratias, A.; Meyers, B.C.; Geffroy, V. Molecular mechanisms that limit the costs of NLR-mediated resistance in plants. Mol. Plant Pathol. 2018, 19, 2516–2523. [Google Scholar] [CrossRef]
- Slootweg, E.J.; Spiridon, L.N.; Roosien, J.; Butterbach, P.; Pomp, R.; Westerhof, L.; Wilbers, R.; Bakker, E.; Bakker, J.; Petrescu, A.-J.; et al. Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 2013, 162, 1510–1528. [Google Scholar] [CrossRef]
- De Oliveira, A.S.; Koolhaas, I.; Boiteux, L.S.; Caldararu, O.F.; Petrescu, A.J.; Oliveira Resende, R.; Kormelink, R. Cell death triggering and effector recognition by Sw-5 SD-CNL proteins from resistant and susceptible tomato isolines to Tomato spotted wilt virus. Mol. Plant Pathol. 2016, 17, 1442–1454. [Google Scholar] [CrossRef]
- Kim, S.B.; Lee, H.Y.; Choi, E.H.; Park, E.; Kim, J.H.; Moon, K.B.; Kim, H.S.; Choi, D. The coiled-coil and leucine-rich repeat domain of the potyvirus resistance protein Pvr4 has a distinct role in signaling and pathogen recognition. Mol. Plant Microbe Interact. 2018, 31, 906–913. [Google Scholar] [CrossRef]
- Tameling, W.I.L.; Vossen, J.H.; Albrecht, M.; Lengauer, T.; Berden, J.A.; Haring, M.A.; Cornelissen, B.J.C.; Takken, F.L.W. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006, 140, 1233–1245. [Google Scholar] [CrossRef]
- Williams, S.J.; Sornaraj, P.; deCourcy-Ireland, E.; Menz, R.I.; Kobe, B.; Ellis, J.G.; Dodds, P.N.; Anderson, P.A. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 2011, 24, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Tian, A.; Miyashita, S.; Ando, S.; Takahashi, H. Single amino acid substitutions in the cucumber mosaic virus 1a protein induce necrotic cell death in virus-inoculated leaves without affecting virus multiplication. Viruses 2020, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Julio, E.; Candresse, T.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Moury, B.; Glais, L.; Dorlhac de Borne, F.; Decroocq, V.; et al. NtTPN1: A RPP8-like R gene required for potato virus Y-induced veinal necrosis in tobacco. Plant J. 2018, 95, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Abebe, D.A.; van Bentum, S.; Suzuki, M.; Ando, S.; Takahashi, H.; Miyashita, S. Plant death caused by inefficient induction of antiviral R-gene-mediated resistance may function as a suicidal population resistance mechanism. Commun. Biol. 2021, 4, 947. [Google Scholar] [CrossRef]
- Pagán, I.; Garcia-Arenal, F. Cucumber mosaic virus-induced systemic necrosis in Arabidopsis thaliana: Determinants and role in plant defense. Viruses 2022, 14, 2790. [Google Scholar] [CrossRef]
- Lukan, T.; Baebler, Š.; Pompe-Novak, M.; Guček, K.; Zagorščak, M.; Coll, A.; Gruden, K. Cell death is not sufficient for the restriction of potato virus Y spread in hypersensitive response-conferred resistance in potato. Front. Plant Sci. 2018, 9, 168. [Google Scholar] [CrossRef]
- Sedlar, A.; Gerič Stare, B.; Mavrič Pleško, I.; Dolničar, P.; Maras, M.; Šuštar-Vozlič, J.; Baebler, Š.; Gruden, K.; Meglič, V. Expression and regulation of programmed cell death-associated genes in systemic necrosis of PVYNTN susceptible potato tubers. Plant Pathol. 2018, 67, 1238–1252. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef]
- Martin, I.R.; Vigne, E.; Velt, A.; Hily, J.-M.; Garcia, S.; Baltenweck, R.; Komar, V.; Rustenholz, C.; Hugueney, P.; Lemaire, O.; et al. Severe stunting symptoms upon nepovirus infection are reminiscent of a chronic hypersensitive-like response in a perennial woody fruit crop. Viruses 2021, 13, 2138. [Google Scholar] [CrossRef]
- Király, L.; Albert, R.; Zsemberi, O.; Schwarczinger, I.; Hafez, Y.M.; Künstler, A. Reactive oxygen species contribute to symptomless, extreme resistance to potato virus x in tobacco. Phytopathology 2021, 111, 1870–1884. [Google Scholar] [CrossRef]
- Sukarta, O.C.A.; Zheng, Q.; Slootweg, E.J.; Mekken, M.; Mendel, M.; Putker, V.; Bertran, A.; Brand, A.; Overmars, H.; Pomp, R.; et al. GLYCINE-RICH RNA-BINDING PROTEIN 7 potentiates effector-triggered immunity through an RNA recognition motif. Plant Physiol. 2022, 189, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Ross, B.T.; Zidack, N.K.; Flenniken, M.L. Extreme resistance to viruses in potato and soybean. Front. Plant Sci. 2021, 12, 658981. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, A.; Takken, F.L.W.; Sánchez-Camargo, V.A. Translation arrest: A key player in plant antiviral response. Genes 2023, 14, 1293. [Google Scholar] [CrossRef] [PubMed]
- Divéki, Z.; Salánki, K.; Balázs, E. The necrotic pathotype of the cucumber mosaic virus (CMV) Ns strain is solely determined by amino acid 461 of the 1a protein. Mol. Plant Microbe Interact. 2004, 17, 837–845. [Google Scholar] [CrossRef]
- Osterbaan, L.J.; Choi, J.; Kenney, J.; Flasco, M.; Vigne, E.; Schmitt-Keichinger, C.; Rebelo, A.R.; Heck, M.; Fuchs, M. The identity of a single residue of the RNA-dependent RNA polymerase of grapevine fanleaf virus modulates vein clearing symptoms in Nicotiana benthamiana. Mol. Plant Microbe Interact. 2019, 32, 790–801. [Google Scholar] [CrossRef]
- Fujita, N.; Komatsu, K.; Ayukawa, Y.; Matsuo, Y.; Hashimoto, M.; Netsu, O.; Teraoka, T.; Yamaji, Y.; Namba, S.; Arie, T. N-terminal region of cysteine-rich protein (CRP) in carlaviruses is involved in the determination of symptom types. Mol. Plant Pathol. 2018, 19, 180–190. [Google Scholar] [CrossRef]
- Hashimoto, M.; Komatsu, K.; Iwai, R.; Keima, T.; Maejima, K.; Shiraishi, T.; Ishikawa, K.; Yoshida, T.; Kitazawa, Y.; Okano, Y.; et al. Cell death triggered by a putative amphipathic helix of radish mosaic virus helicase protein is tightly correlated with host membrane modification. Mol. Plant Microbe Interact. 2015, 28, 675–688. [Google Scholar] [CrossRef]
- Kuroiwa, M.; Handa, S.; Gyoutoku, Y.; Moriyama, M.; Neriya, Y.; Nishigawa, H.; Natsuaki, T. Characterization of a ToMV isolate overcoming Tm-22 resistance gene in tomato. Virus Genes 2022, 58, 478–482. [Google Scholar] [CrossRef]
- Liu, J.; Wu, X.; Fang, Y.; Liu, Y.; Bello, E.O.; Li, Y.; Xiong, R.; Li, Y.; Fu, Z.Q.; Wang, A.; et al. A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity. Nat. Commun. 2023, 14, 3580. [Google Scholar] [CrossRef]
- Zhou, J.M.; Zhang, Y. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ding, P.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.-W.; Zhou, J.-M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, 44. [Google Scholar] [CrossRef] [PubMed]
- Bi, G.; Su, M.; Li, N.; Liang, Y.; Dang, S.; Xu, J.; Hu, M.; Wang, J.; Zou, M.; Deng, Y.; et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 2021, 184, 3528–3541. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lapin, D.; Liu, L.; Sun, Y.; Song, W.; Zhang, X.; Logemann, E.; Yu, D.; Wang, J.; Jirschitzka, J.; et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 2020, 370, 1184. [Google Scholar] [CrossRef]
- Martin, R.; Qi, T.; Zhang, H.; Liu, F.; King, M.; Toth, C.; Nogales, E.; Staskawicz, B.J. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 2020, 370, 1185. [Google Scholar] [CrossRef]
- Mestre, P.; Baulcombe, D.C. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 2006, 18, 491–501. [Google Scholar] [CrossRef]
- Jacob, P.; Kim, N.H.; Wu, F.; El-Kasmi, F.; Chi, Y.; Walton, W.G.; Furzer, O.J.; Lietzan, A.D.; Sunil, S.; Kempthorn, K.; et al. Plant “helper” immune receptors are Ca(2+)-permeable nonselective cation channels. Science 2021, 373, 420–425. [Google Scholar] [CrossRef]
- Feehan, J.M.; Wang, J.; Sun, X.; Choi, J.; Ahn, H.K.; Ngou, B.P.M.; Parker, J.E.; Jones, J.D.G. Oligomerization of a plant helper NLR requires cell-surface and intracellular immune receptor activation. Proc. Natl. Acad. Sci. USA 2023, 120, e2210406120. [Google Scholar] [CrossRef]
- Wang, J.; Chen, T.; Han, M.; Qian, L.; Li, J.; Wu, M.; Han, T.; Cao, J.; Nagalakshmi, U.; Rathjen, J.P.; et al. Plant NLR immune receptor Tm-22 activation requires NB-ARC domain-mediated self-association of CC domain. PLoS Pathog. 2020, 16, e1008475. [Google Scholar] [CrossRef]
- Huang, S.; Jia, A.; Ma, S.; Sun, Y.; Chang, X.; Han, Z.; Chai, J. NLR signaling in plants: From resistosomes to second messengers. Trends Biochem. Sci. 2023, 48, 776–787. [Google Scholar] [CrossRef]
- Castel, B.; Ngou, P.-M.; Cevik, V.; Redkar, A.; Kim, D.-S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Seong, K.; Thomazella, D.P.T.; Kim, J.R.; Pham, J.; Seo, E.; Cho, M.-J.; Schultink, A.; Staskawicz, B.J. NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2018, 115, E10979–E10987. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Derevnina, L.; Kamoun, S. Receptor networks underpin plant immunity. Science 2018, 360, 1300–1301. [Google Scholar] [CrossRef] [PubMed]
- Lüdke, D.; Yan, Q.; Rohmann, P.F.W.; Wiermer, M. NLR we there yet? Nucleocytoplasmic coordination of NLR-mediated immunity. New Phytol. 2022, 236, 24–42. [Google Scholar] [CrossRef]
- Bhandari, D.D.; Lapin, D.; Kracher, B.; von Born, P.; Bautor, J.; Niefind, K.; Parker, J.E. An EDS1 heterodimer signalling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat. Commun. 2019, 10, 772. [Google Scholar] [CrossRef]
- Bernoux, M.; Chen, J.; Zhang, X.; Newell, K.; Hu, J.; Deslandes, L.; Dodds, P. Subcellular localization requirements and specificities for plant immune receptor Toll-interleukin-1 receptor signaling. Plant J. 2023, 114, 1319–1337. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, Q.; Tong, C.; Chen, H.; Miao, D.; Qian, X.; Zhao, X.; Jiang, L.; Tao, X. Characterization of the roles of SGT1/RAR1, EDS1/NDR1, NPR1, and NRC/ADR1/NRG1 in Sw-5b-mediated resistance to tomato spotted wilt virus. Viruses 2021, 13, 1447. [Google Scholar] [CrossRef]
- Ding, P.; Ding, Y. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Knip, M.; Schachtschabel, J.; Beijaert, M.S.; Takken, F.L.W. Perturbation of nuclear-cytosolic shuttling of Rx1 compromises extreme resistance and translational arrest of potato virus X transcripts. Plant J. 2021, 106, 468–479. [Google Scholar] [CrossRef]
- Machado, J.P.B.; Calil, I.P.; Santos, A.A.; Fontes, E.P.B. Translational control in plant antiviral immunity. Genet. Mol. Biol. 2017, 40, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Hoser, R.; Żurczak, M.; Lichocka, M.; Zuzga, S.; Dadlez, M.; Samuel, M.A.; Ellis, B.E.; Stuttmann, J.; Parker, J.E.; Hennig, J.; et al. Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol. 2013, 200, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Tameling, W.I.; Nooijen, C.; Ludwig, N.; Boter, M.; Slootweg, E.; Goverse, A.; Shirasu, K.; Joosten, M.H. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 2010, 22, 4176–4194. [Google Scholar] [CrossRef]
- Townsend, P.D.; Dixon, C.H.; Slootweg, E.J.; Sukarta, O.C.A.; Yang, A.W.H.; Hughes, T.R.; Sharples, G.J.; Palsson, L.O.; Takken, F.L.W.; Goverse, A.; et al. The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. J. Biol. Chem. 2018, 293, 3218–3233. [Google Scholar] [CrossRef] [PubMed]
- Sukarta, O.C.A.; Townsend, P.D.; Llewelyn, A.; Dixon, C.H.; Slootweg, E.J.; Pålsson, L.O.; Takken, F.L.W.; Goverse, A.; Cann, M.J. A DNA-binding bromodomain-containing protein interacts with and reduces Rx1-mediated immune response to potato virus X. Plant Commun. 2020, 1, 100086. [Google Scholar] [CrossRef]
- Chen, H.; Qian, X.; Chen, X.; Yang, T.; Feng, M.; Chen, J.; Cheng, R.; Hong, H.; Zheng, Y.; Mei, Y.; et al. Cytoplasmic and nuclear Sw-5b NLR act both independently and synergistically to confer full host defense against tospovirus infection. New Phytol. 2021, 231, 2262–2281. [Google Scholar] [CrossRef]
- Sharma, N.; Prasad, A.; Prasad, M. Role of the Sw5 gene cluster in the fight against plant viruses. J. Virol. 2022, 96, e0208421. [Google Scholar] [CrossRef]
- Qin, Y.; Zhao, J.; Wang, J.; Ye, X.; Zhou, C.; Zhou, Y. Regulation of Nicotiana benthamiana cell death induced by citrus chlorotic dwarf-associated virus-RepA protein by WRKY 1. Front. Plant Sci. 2023, 14, 1164416. [Google Scholar] [CrossRef]
- Xu, Y.-P.; Zhao, Y.; Song, X.-Y.; Ye, Y.-F.; Wang, R.-G.; Wang, Z.-L.; Ren, X.-L.; Cai, X.-Z. Ubiquitin extension protein UEP1 modulates cell death and resistance to various pathogens in tobacco. Phytopathology 2019, 109, 1257–1269. [Google Scholar] [CrossRef]
- Kumar, S.; Zavaliev, R.; Wu, Q.; Zhou, Y.; Cheng, J.; Dillard, L.; Powers, J.; Withers, J.; Zhao, J.; Guan, Z.; et al. Structural basis of NPR1 in activating plant immunity. Nature 2022, 605, 561–566. [Google Scholar] [CrossRef]
- Zavaliev, R.; Mohan, R.; Chen, T.; Dong, X. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 2020, 182, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Park, S.H.; Chua, N.H. UBP12/UBP13-mediated deubiquitination of salicylic acid receptor NPR3 suppresses plant immunity. Mol. Plant 2023, 16, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Li, G.; Liu, M.; Liu, R.; Yang, S.; Wang, K.; Lu, L.; Ye, Q.; Liu, J.; Liang, J.; et al. A ubiquitin-specific protease functions in regulating cell death and immune responses in rice. Plant Cell Environ. 2023, 46, 1312–1326. [Google Scholar] [CrossRef] [PubMed]
- Sertsuvalkul, N.; DeMell, A.; Dinesh-Kumar, S.P. The complex roles of autophagy in plant immunity. FEBS Lett. 2022, 596, 2163–2171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hong, Q.; Li, Y.; Li, Q.; Wang, M. Autophagy contributes to regulate the ROS levels and PCD progress in TMV-infected tomatoes. Plant Sci. 2018, 269, 12–19. [Google Scholar] [CrossRef]
- Han, S.; Wang, Y.; Zheng, X.; Jia, Q.; Zhao, J.; Bai, F.; Hong, Y.; Liu, Y. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 2015, 27, 1316–1331. [Google Scholar] [CrossRef]
- Jeon, H.S.; Jang, E.; Kim, J.; Kim, S.H.; Lee, M.H.; Nam, M.H.; Tobimatsu, Y.; Park, O.K. Pathogen-induced autophagy regulates monolignol transport and lignin formation in plant immunity. Autophagy 2023, 19, 597–615. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, S.; Yang, X.; Yang, X.; Zhang, T.; Zhou, G. Friend or enemy: A dual role of autophagy in plant virus infection. Front. Microbiol. 2020, 11, 736. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Wang, Y.; Zheng, X.; Wei, X.; Nagalakshmi, U.; et al. Cotton leaf curl Multan virus βC1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell 2020, 32, 1124–1135. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, L.; Li, L.; Zhou, T.; Yan, F.; Zhang, H.; Zhu, Y.; Andika, I.B.; Sun, L. Coat protein of rice stripe virus enhances autophagy activity through interaction with cytosolic glyceraldehyde-3-phosphate dehydrogenases, a negative regulator of plant autophagy. Stress Biol. 2023, 3, 3. [Google Scholar] [CrossRef]
- Niu, E.; Ye, C.; Zhao, W.; Kondo, H.; Wu, Y.; Chen, J.; Andika, I.B.; Sun, L. Coat protein of Chinese wheat mosaic virus upregulates and interacts with cytosolic glyceraldehyde-3-phosphate dehydrogenase, a negative regulator of plant autophagy, to promote virus infection. J. Integr. Plant Biol. 2022, 64, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ismayil, A.; Jiang, Z.; Wang, Y.; Zheng, X.; Yan, L.; Hong, Y.; Li, D.; Liu, Y. A viral protein disrupts vacuolar acidification to facilitate virus infection in plants. EMBO J. 2022, 41, e108713. [Google Scholar] [CrossRef]
- O’Leary, B.M. The case of virus-induced plant autophagy: Cui bono? Plant Cell 2020, 32, 805–806. [Google Scholar] [CrossRef] [PubMed]
- Salguero-Linares, J.; Coll, N.S. Plant proteases in the control of the hypersensitive response. J. Exp. Bot. 2019, 70, 2087–2095. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef]
- Cesari, S.; Xi, Y.; Declerck, N.; Chalvon, V.; Mammri, L.; Pugnière, M.; Henriquet, C.; de Guillen, K.; Chochois, V.; Padilla, A.; et al. New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 2022, 13, 1524. [Google Scholar] [CrossRef]
- Kourelis, J.; Marchal, C.; Posbeyikian, A.; Harant, A.; Kamoun, S. NLR immune receptor–nanobody fusions confer plant disease resistance. Science 2023, 379, 934–939. [Google Scholar] [CrossRef]
- Tran, P.T.; Widyasari, K.; Park, J.Y.; Kim, K.H. Engineering an auto-activated R protein that is in vivo activated by a viral protease. Virology 2017, 510, 242–247. [Google Scholar] [CrossRef]
- Contreras, M.P.; Pai, H.; Selvaraj, M.; Toghani, A.; Lawson, D.M.; Tumtas, Y.; Duggan, C.; Yuen, E.L.H.; Stevenson, C.E.M.; Harant, A.; et al. Resurrection of plant disease resistance proteins via helper NLR bioengineering. Sci. Adv. 2023, 9, eadg3861. [Google Scholar] [CrossRef]


| Year | ER/HR | Plant of Resistance Origine | Resistance Gene(s)/Locus | Type of NLR | Virus (Genus; Family) | Viral Determinant | References | Cloning Strategy | Identification of Cognate Avr | Confirmation of R Function |
|---|---|---|---|---|---|---|---|---|---|---|
| 1995 | HR | Tobacco Nicotiana glutinosa | N | TIR-NBS-LRR | Tobacco mosaic virus = TMV (Tobamovirus; Virgaviridae) | 130K Replicase (p50 helicase domain) | [50] [78] | Transposon tagging | Chimeric viruses | Transgenic expression |
| 1999 | ER (SHR in transgenic N. benthamiana) | Wild potato Solanum andigena | Rx-1 (Chr XII) | CC-NBS-LRR | Potato virus X = PVX (Potexvirus; Alfaflexiviridae) | CP | [79] [80] | Map-based cloning | Chimeric viruses (TMV vector expressing PVX CP) | Transgenic expression |
| 2000 | ER (HR under weak promoter in tobacco) | Potato Solanum acuale | Rx-2 (Chr V) | CC-NBS-LRR | PVX (Potexvirus; Alfaflexiviridae | CP | [71] [81] | Agrobacterium-mediated expression of a library of Rx-homologues | Point mutations in CP gene | Transgenic expression |
| 2000 | HR | Thale cress Arabidopsis thaliana | HRT | CC-NBS-LRR | Turnip crinkle virus = TCV (Betacarmovirus; Tombusviridae | CP | [82] | Map-based cloning | Transgenic expression | Transgenic expression |
| 2001 | ER | Tomato Solanum peruvianum | Sw5-b | CC-NBS-LRR | Tomato spotted wilt virus = TSWV and other tospoviruses (Orthotospovirus; Tospoviridae) | Nsm (MP) | [83] [84] | Map-based cloning Bac screening | Chimeric viruses (AlMV vector with MP of TSWV) | Transgenic expression |
| 2002 | HR | Thale cress Arabidopsis thaliana | RCY1 same locus as HRT | CC-NBS-LRR | Cucumber mosaic virus = CMV (Cucumovirus; Bromoviridae) | CP | [85] [86] | Map-based cloning | Chimeric viruses | Transgenic expression |
| 2002 | ER to PVA & PVV? Systemic necrosis to PVY | Potato Solanum tuberosum | Y-1 (Chr XI) | TIR-NBS-LRR | Potato virus Y = PVY, potato virus A = PVA, potato virus V = PVV (Potyvirus; Potyviridae) | ? | [87] | Homology cloning | - | Transgenic expression |
| 2003 | ER | Tomato Solanum peruvianum | Tm-2 (alleles Tm-2 & Tm-22) | CC-NBS-LRR | Tomato mosaic virus = ToMV & TMV (Tobamovirus; Virgaviridae) | 30 K MP (C-term) | [67] [88] | Transposon tagging Homology cloning (Tm-22) | Deletion of C-terminus of MP in virus Transgenic expression | Transgenic expression |
| 2006 | SHR in transgenic N. benthamiana (ER in P. vulgaris?) | Common bean Phaseolus vulgaris | PvCMR1 (RT4-4) | TIR-NBS-LRR | CMV (Cucumovirus; Bromoviridae) | 2a | [89] | Homology cloning | Transient expression (agrobacterium) | Transgenic expression |
| 2007 | HR | Common bean Phaseolus vuulgaris | PvVTT1 | TIR-NBS-LRR | Bean dwarf mosaic virus = BDMV (Begomovirus; Geminiviridae) | BV1 (NSP) | [90] [91] | cDNA substraction & cloning | Chimeric viruses | Transgenic expression |
| 2010 | HR | Chinese cabbage Brassica campestric | BcTuR3 (cloned but not confirmed) | TIR-NBS-LRR | Turnip mosaic virus = TuMV (Potyvirus; Potyviridae) | ? | [92] | Homology cloning | - | Not confirmed |
| 2011 | HR | Pepper Capsicum spp. | L (alleles L2–L4) | CC-NBS-LRR | TMV, ToMV, paprika mild mottle virus = PaMMV, peper milds mottle virus = PMMV (Tobamovirus; Virgaviridae) | CP | [74] [93] | Map-based (L3) andhomology-based cloning (L1, L2, L4) | Chimeric viruses & mutants | Transient expression (agrobacterium) |
| 2012 | HR | White tobacco Nicotiana sylvestris | N′ | CC-NBS-LRR | TMV & other tobamoviruses (Tobamovirus; Virgaviridae) | CP | [75] [94] | Homology cloning (L) | Point mutations in CP gene | Transient expression (agrobacterium) |
| 2012 | HR | Black gram Vigna mungo | CYR1 | CC-NBS-LRR | Mungbean yellow mosaic India virus = MYMIV (Begomovirus; Geminiviridae) | AV1 (CP)? | [95] | Map-based cloning | In silico model | Not confirmed |
| 2013 | ER | Melon Cucumis melo | Prv | TIR-NBS-LRR | Papaya ringspot virus = PRSV (Potyvirus; Potyviridae) | ? | [96] [97] | Map-based cloning BAC screening | - | Genome editing |
| 2014 | HR | Pepper | Pvr9 (Chr VI) | CC-NBS-LRR | Pepper mottle virus = PepMoV (Potyvirus; Potyviridae) | NIb (Pol) | [72] [73] | Agrobacterium-mediated expression of a library of R candidates | Transient expression (agrobacterium) | Transient expression (agrobacterium) |
| 2014 | ER | Turnip Brassica rapa | TuRBO7 (1 candidate gene, not identified, not cloned) | CC-NBS-LRR | TuMV (Potyvirus; Potyviridae) | ? | [98] | Map-based cloning | - | Not confirmed |
| 2016 | ER (SHR with strain SMV-G7) | Soybean Glycine max | Rsv1-h (2 candidate genes identified, not cloned) | CC-NBS-LRR | Soybean mosaic virus = SMV (Potyvirus; Potyviridae) | P3 & HC-Pro | [99] [100] | Map-based cloning | Chimeric viruses & mutants | Not confirmed |
| 2017 | ER / HR | Pepper Capsicum annuum | Pvr4 / Pvr7 (ChrX) | CC-NBS-LRR | PepMoV, PVY (Potyvirus; Potyviridae) | NIb (Pol) | [76] [101] [102] | Map-based cloning BAC screening | Transient expression (agrobacterium) | Transient expression (agrobacterium) |
| 2017 | HR | Pepper Capsicum annuum | Tsw (Chr X) same locus as Pvr4 | CC-NBS-LRR | TSWV (Orthotospovirus; Tospoviridae) | NSs (VSR) | [76] [103] | Map-based cloning BAC screening | Transient expression (agrobacterium) | Transient expression (agrobacterium) |
| 2017 | HR | Sugar beet Beta vulgaris | Rz2 | CC-NBS-LRR | Beet necrotic yellow vein virus = BNYVV (Benyvirus; Benyviridae) | TGB1 (MP) | [104] [105] | Mapping-by-sequuencing | Transient expression (agrobacterium) | Silencing by hairpin in transgenic sugarbeet |
| 2018 | ER / HR | Wild tomato Solanum habrochaites | Ty-2 = TYNBS1 | CC-NBS-LRR | Tomato yellow leaf curl virus = TYLCV (Begomovirus; Geminiviridae) | Rep / C1 (replication-associated protein) | [77] [106] | Map-based cloning and transgenic expression | Transient expression (agrobacterium) | Transgenic expression |
| 2020 | ER | Wild potato Solanum stoloniferum | Rysto = Ry-fsto | TIR-NBS-LRR | PVY, PVA (Potyvirus; Potyviridae) | CP / previously NIa involved | [66] [107] | Enrichment sequencing & Pac Bio single-molecule real-time sequencing (SMRT RenSeq) | Transient expression (agrobacterium) | Transient expression (agrobacterium) |
| 2021 | HR | Tomato Solanum lycopersicum | Sw5-a | CC-NBS-LRR | Tomato leaf curl New Dehli virus = ToLCNDV (Begomovirus; Geminiviridae) | AC4 (VSR, suppressor of cell death) | [108] | miRNAomics identified a regulator of Sw5-a | Identification of Sw5-a interactant Transient expression (agrobacterium) | Virus induced gene silencing |
| 2021 | ER | Soybean Glycine max | Rsc 4-3 = Rsv 3 / NBS_C | CC-NBS-LRR | SMV (Potyvirus; Potyviridae) | CI (cylindrical inclusion protein, replication & movement) | [70] | Fine mapping Transient expression (agrobacterium) Cas9-assisted mutation of candidate genes | Chimeric viruses & transient expression (agrobacterium) | Transient expression (agrobacterium) & genome editing |
| 2022 | ER | Wild potato Solanum chacosense | Rychc (ChriX) | TIR-NBS-LRR | PVY (Potyvirus; Potyviridae) | ? | [109] | Map-based cloning | - | Transgenic expression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piau, M.; Schmitt-Keichinger, C. The Hypersensitive Response to Plant Viruses. Viruses 2023, 15, 2000. https://doi.org/10.3390/v15102000
Piau M, Schmitt-Keichinger C. The Hypersensitive Response to Plant Viruses. Viruses. 2023; 15(10):2000. https://doi.org/10.3390/v15102000
Chicago/Turabian StylePiau, Maïlys, and Corinne Schmitt-Keichinger. 2023. "The Hypersensitive Response to Plant Viruses" Viruses 15, no. 10: 2000. https://doi.org/10.3390/v15102000
APA StylePiau, M., & Schmitt-Keichinger, C. (2023). The Hypersensitive Response to Plant Viruses. Viruses, 15(10), 2000. https://doi.org/10.3390/v15102000







