Development of Polyclonal Antibodies and a Serological-Based Reverse-Transcription Loop-Mediated Isothermal Amplification (S-RT-LAMP) Assay for Rice Black-Streaked Dwarf Virus Detection in Both Rice and Small Brown Planthopper
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction of the Vector for Recombinant CPRBSDV Protein Expression
2.2. Prokaryotic Expression and Purification of the Recombinant His-CPRBSDV Protein
2.3. Production of the Polyclonal Antibody against the His-CPRBSDV Protein
2.4. RT-PCR Detection of Two Rice Viruses
2.5. Three Detection Methods of the RBSDV with Prepared PAb-CPRBSDV
2.6. S-RT-LAMP Detection of the RBSDV with Prepared PAb-CPRBSDV
3. Results
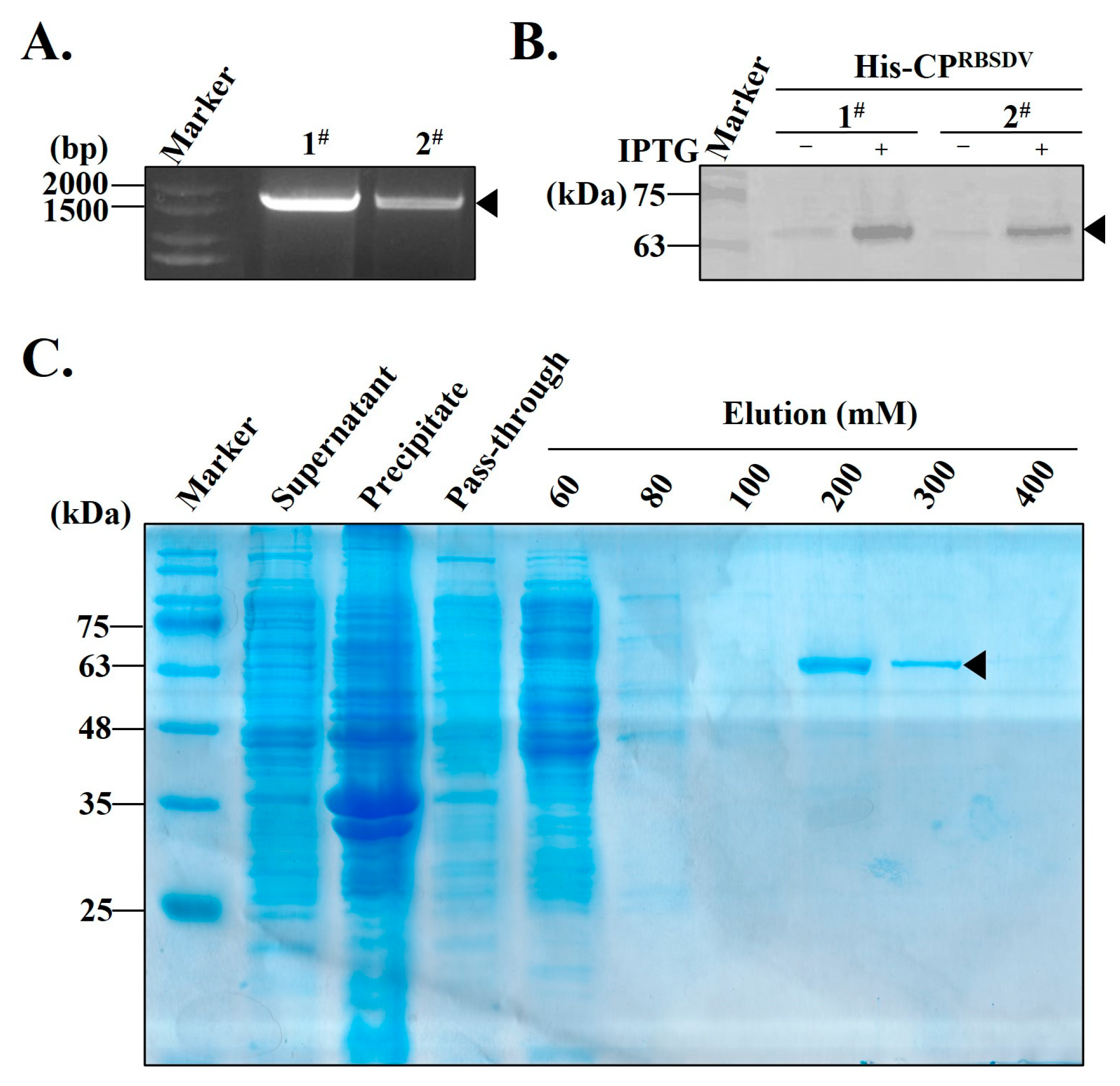
3.1. Expression and Purification of the Recombinant His-CPRBSDV Protein
3.2. Antiserum Production Using the Recombinant His-CPRBSDV Protein
3.3. RT-PCR, Western Blot and Dot Blot Detection of RBSDV Infection in Rice Plants or SBPHs Using the Prepared PAb-CPRBSDV
3.4. Establishment of Specific ELISA Detection Methods
3.5. Establishment of S-RT-LAMP Detection Method Based on Serological-Based and RT-LAMP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keiichiro, M.; Sachiyo, S.-M.; Tomohisa, F.; Masaya, M. Potential risks of poaceous plants as infectious sources of Rice black-streaked dwarf virus transmitted by the small brown planthopper, Laodelphax striatellus. Plant Dis. 2019, 103, 1244–1248. [Google Scholar]
- Wu, N.; Zhang, L.; Ren, Y.; Wang, X. Rice black-streaked dwarf virus: From multiparty interactions among plant-virus-vector to intermittent epidemics. Mol. Plant Pathol. 2020, 21, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, X.; Chen, J.; Sun, L. A phloem-limited fijivirus induces the formation of neoplastic phloem tissues that house virus multiplication in the host plant. Sci. Rep. 2016, 6, 29848. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Peck, X.Y.; Choong, Y.K.; Ng, W.S.; Engl, W.; Raghuvamsi, P.V.; Zhao, Z.W.; Anand, G.S.; Zhou, Y.; Sivaraman, J.; et al. Identification of putative binding interface of PI(3,5)P(2) lipid on rice black-streaked dwarf virus (RBSDV) P10 protein. Virology 2022, 570, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zhu, J.; Feng, J.; Tao, X.; Zhong, X.-M.; Sun, X. Study on the relationship between dwarf level and yield in rices infected with black-streaked dwarf disease. Acta Agric. Zhejiangensis 2013, 25, 298–302. [Google Scholar]
- Zhang, L.; Wu, N.; Ren, Y.; Wang, X. Insights into insect vector transmission and epidemiology of plant-infecting Fijiviruses. Front. Microbiol. 2021, 12, 628262. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Z.; Huang, C.; Lu, M.; Liu, J.; Yang, Q. Statistics and analysis of crop yield losses caused by main diseases and insect pests in recent 10 years. Plant Prot. 2016, 42, 1–9. [Google Scholar]
- Zhang, H.-M.; Lei, J.-L.; Chen, J.-P.; Lu, Y.-P.; Chen, S.-X.; Xue, Q.-Z.; Adams, M.J. A dwarf disease on rice, wheat and maize from Zhejiang and Hebei is caused by Rice black-streaked dwarf virus. Virol. Sin. 2001, 16, 246–251. [Google Scholar]
- Zhang, H.M.; Chen, J.P.; Lei, J.L.; Adams, M.J. Sequence analysis shows that a dwarfing disease on rice, wheat and maize in China is caused by Rice black-streaked dwarf virus. Eur. J. Plant Pathol. 2001, 107, 563–567. [Google Scholar] [CrossRef]
- Isogai, M.; Uyeda, I.; Lee, B.-C. Detection and assignment of proteins encoded by Rice black streaked dwarf fijivirus S7, S8, S9 and S10. J. Gen. Virol. 1998, 79, 1487–1494. [Google Scholar] [CrossRef]
- Liu, H.; Wei, C.; Zhong, Y.; Li, Y. Rice black-streaked dwarf virus outer capsid protein P10 has self-interactions and forms oligomeric complexes in solution. Virus Res. 2007, 127, 34–42. [Google Scholar] [CrossRef]
- Wu, J.; Ni, Y.; Liu, H.; Rao, L.; Zhou, Y.; Zhou, X. Development and use of three monoclonal antibodies for the detection of Rrice black-streaked dwarf virus in field plants and planthopper vectors. Virol. J. 2013, 10, 114. [Google Scholar] [CrossRef]
- Azuhata, F.; Uyeda, I.; Kimura, I.; Shikata, E. Close similarity between genome structures of rice black-streaked dwarf and maize rough dwarf viruses. J. Gen. Virol. 1993, 74 Pt 7, 1227–1232. [Google Scholar] [CrossRef]
- Wang, Z.-H.; Fang, S.-G.; Zhang, Z.-Y.; Han, C.-G.; Li, D.-W.; Yu, H.-L. Development of an ID-ELISA for the detection of Rice black-streaked dwarf virus in plants. J. Virol. Methods 2006, 134, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, Z.; Fan, Y.; Zhou, Y.; Cheng, Z.; Zhang, W. Detection of pathogen of Maize rough dwarf disease (MRDD) in Jiangsu Province with RT-PCR. Nong Ye Sheng Wu Ji Shu Xue Bao 2000, 8, 369–372. [Google Scholar]
- Zhou, T.; Du, L.; Fan, Y.; Zhou, Y. Reverse transcription loop-mediated isothermal amplification of RNA for sensitive and rapid detection of Southern rice black-streaked dwarf virus. J. Virol. Methods 2012, 180, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, H.; Yuan, P.; Zhang, X.; Chen, Q.; Jiang, X.; Zhou, Y. Development of a simplified RT-PCR without RNA isolation for rapid detection of RNA viruses in a single small brown planthopper (Laodelphax striatellus Fallen). Virol. J. 2017, 14, 90. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Xu, H.; Guo, X.; He, Z.; Xu, K.; Liu, F. Sensitive and high-throughput polyclonal antibody-based serological methods for Rice stripe virus detection in both rice and small brown planthopper. Crop Prot. 2021, 144, 105599. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Xu, H.; Gan, H.; He, Z.; Chen, J. Development of polyclonal antibodies-based serological methods and a DIG-labelled DNA probe-based molecular method for detection of the Vicia cryptic virus-M in field plants. J. Virol. Methods 2022, 299, 114331. [Google Scholar] [CrossRef]
- Chen, J.; Feng, C.; Guo, X.; Zhou, Y.; Gu, T.; Zhuang, X.; Cheng, L.; Zhang, K. Development of polyclonal antibodies-based serological methods for detection of the Rehmannia mosaic virus in field plants. Front. Sustain. Food Syst. 2022, 6, 1013470. [Google Scholar] [CrossRef]
- Wang, G.; Zhou, Y.; Chen, Z.; Zhou, X. Production of monoclonal antibodies to rice stripe virus and application in virus detection. Acta Phytopathol. Sin. 2004, 34, 302–306. [Google Scholar]
- Boccardo, G.; Milne, R.G. Enhancement of the immunogenicity of the Maize rough dwarf virus outer shell with the cross-linking reagent dithiobis (succinimidyl) propionate. J. Virol. Methods 1981, 3, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Shikata, E.; Kitagawa, Y. Rice black-streaked dwarf virus: Its properties, morphology and intracellular localization. Virology 1977, 77, 826–842. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, H.M.; Ying, L.; Li, J.; Lv, M.F.; Xie, L.; Li, P.P.; Liu, X.Y.; Liang-Ying, D.; Chen, J.P. Rice black-streaked dwarf virus genome segment S5 is a bicistronic mRNA in infected plants. Arch. Virol. 2014, 159, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Festus, R.O.; Seal, S.E.; Prempeh, R.; Quain, M.D.; Silva, G. Improved reverse transcription loop-mediated isothermal amplification (RT-LAMP) for the rapid and sensitive detection of Yam mosaic virus. Viruses 2023, 15, 1592. [Google Scholar] [CrossRef]
- Madhu Kovileri, M.; Nair, S.; Loius, V. One-step Reverse Transcription-loop mediated isothermal amplification (RT-LAMP) for closed-tube colorimetric detection of Banana bract mosaic virus in Banana (Musa spp.). 3 Biotech 2023, 13, 131. [Google Scholar] [CrossRef]
- Zhang, P.; Mar, T.T.; Liu, W.; Li, L.; Wang, X. Simultaneous detection and differentiation of Rice black streaked dwarf virus (RBSDV) and Southern rice black streaked dwarf virus (SRBSDV) by duplex real time RT-PCR. Virol. J. 2013, 10, 24. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, S.; Gao, R.; Sun, F.; Liu, W.; Zhou, G.; Wu, J.; Zhou, X.; Zhou, Y. Distribution and genetic diversity of Southern rice black-streaked dwarf virus in China. Virol. J. 2013, 10, 307. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, J.; Zhou, G.; Zhang, H.; Chen, J.; Adams, M.J. The complete genome sequence of two isolates of Southern rice black-streaked dwarf virus, a new member of the genus Fijivirus. J. Phytopathol. 2010, 158, 733–737. [Google Scholar] [CrossRef]




| Primer Name | Primer Sequences (5′-3′) a | Length of PCR Products (bp) b | Tm Values (°C) | Purpose |
|---|---|---|---|---|
| RBSDV/P10/F | ATGGCTGACATAAGACTCG | 1677 | 53.01 | RT-PCR amplification of the S10 fragment of RBSDV |
| RBSDV/P10/R | CGCACAGCACTGAACTAGTC | 57.45 | ||
| pET28a/P10/EcoR I/F | GGAATTCATGGCTGACATAAGACTCG | 1677 | 59.59 | Construction of the prokaryotic expression vector |
| pET28a/P10/Xho I/R | CCGCTCGAGTCTTGTCACTTTATTTAATAC | 59.63 | ||
| RSV/NSvc3/F | ATGGGCACCAACAAGCCAGC | 981 | 62.23 | RT-PCR amplification of the NSvc3 fragment of RSV |
| RSV/NSvc3/R | GTCATCTGCACCTTCTGCCTC | 59.14 |
| Primer Name | Type Primer | Length of Primers | Genome Position |
|---|---|---|---|
| F3 | Forward outer | 21-mer | 279–299 |
| B3 | Backward outer | 21-mer | 467–447 |
| FIP (F1c + F2) | Forward inner | 45-mer | 366–344, 304–325 |
| BIP (B1c + B2) | Backward inner | 47-mer | 374–398, 444–423 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Feng, C.; Gu, T.; Chen, H.; Liu, D.; Xu, K.; Zhang, K. Development of Polyclonal Antibodies and a Serological-Based Reverse-Transcription Loop-Mediated Isothermal Amplification (S-RT-LAMP) Assay for Rice Black-Streaked Dwarf Virus Detection in Both Rice and Small Brown Planthopper. Viruses 2023, 15, 2127. https://doi.org/10.3390/v15102127
Hua Y, Feng C, Gu T, Chen H, Liu D, Xu K, Zhang K. Development of Polyclonal Antibodies and a Serological-Based Reverse-Transcription Loop-Mediated Isothermal Amplification (S-RT-LAMP) Assay for Rice Black-Streaked Dwarf Virus Detection in Both Rice and Small Brown Planthopper. Viruses. 2023; 15(10):2127. https://doi.org/10.3390/v15102127
Chicago/Turabian StyleHua, Yanhong, Chenwei Feng, Tianxiao Gu, Haoyu Chen, Duxuan Liu, Kai Xu, and Kun Zhang. 2023. "Development of Polyclonal Antibodies and a Serological-Based Reverse-Transcription Loop-Mediated Isothermal Amplification (S-RT-LAMP) Assay for Rice Black-Streaked Dwarf Virus Detection in Both Rice and Small Brown Planthopper" Viruses 15, no. 10: 2127. https://doi.org/10.3390/v15102127
APA StyleHua, Y., Feng, C., Gu, T., Chen, H., Liu, D., Xu, K., & Zhang, K. (2023). Development of Polyclonal Antibodies and a Serological-Based Reverse-Transcription Loop-Mediated Isothermal Amplification (S-RT-LAMP) Assay for Rice Black-Streaked Dwarf Virus Detection in Both Rice and Small Brown Planthopper. Viruses, 15(10), 2127. https://doi.org/10.3390/v15102127







