Circulation of Dengue Virus Serotype 1 Genotype V and Dengue Virus Serotype 2 Genotype III in Tocantins State, Northern Brazil, 2021–2022
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Aspects
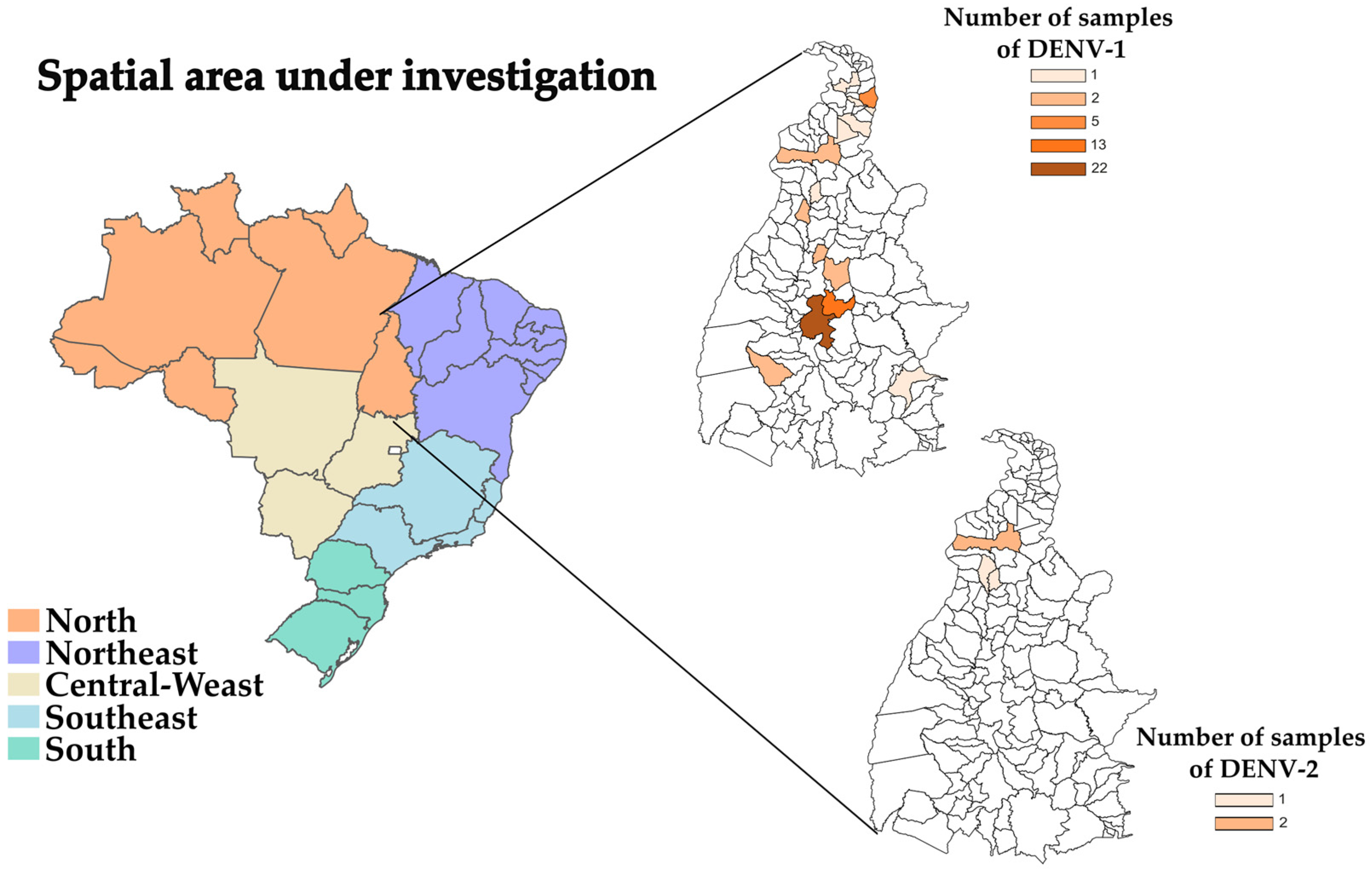
2.2. Sample Collection and RNA Isolation
2.3. Synthesis of cDNA and Multiplex PCR
2.4. Minion Whole Genome Sequencing
2.5. Phylogenetic Analyses
Bayesian Evolutionary Analysis
2.6. Single Nucleotide Polymorphisms (SNPs) Analysis
3. Results
3.1. Phylogenetic Analysis
Bayesian Evolutionary Analysis
3.2. SNPs Analysis
4. Discussion
4.1. Phylogenetic Analysis of DENV-1 and DENV-2
Bayesian Evolutionary Analysis
4.2. SNPs Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siler, J.F.; Hall, M.W.; Hitchens, A. Dengue, its history, epidemiology, mechanism of transmission, etiology, clinical manifestations, immunity and prevention. Philipp J. Sci. 1926, 29, 1–304. [Google Scholar]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Dengue and dengue hemorrhagic fever in the Americas. Puerto Rico Health Sci. J. 1987, 6, 107–111. [Google Scholar]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Lewis, J.G.; Gubler, D.J.; Trent, D.W. Molecular evolution and epidemiology of dengue-3 viruses. J. Gen. Virol. 1994, 75, 65–75. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Gubler, D.J.; Trent, D.W. Molecular evolution and phylogeny of dengue-4 viruses. J. Gen. Virol. 1997, 78, 2279–2286. [Google Scholar] [CrossRef]
- Reich, N.G.; Shrestha, S.; King, A.A.; Rohani, P.; Lessler, J.; Kalayanarooj, S.; Yoon, I.K.; Gibbons, R.V.; Burke, D.S.; Cummings, D.A. Interactions between serotypes of dengue highlight epidemiological impact of cross-immunity. J. R. Soc. Interface 2013, 10, 20130414. [Google Scholar] [CrossRef]
- Sabin, A.B. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1952, 1, 30–50. [Google Scholar] [CrossRef]
- Kyle, J.L.; Harris, E. Global spread and persistence of dengue. Annu. Rev. Microbiol. 2008, 62, 71–92. [Google Scholar] [CrossRef]
- Weaver, S.C.; Vasilakis, N. Molecular evolution of dengue viruses: Contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol. 2009, 9, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Bruycker-Nogueira, F.; Souza, T.M.A.; Chouin-Carneiro, T.; Costa Faria, N.R.; Santos, J.B.; Torres, M.C.; Ramalho, I.L.C.; Aguiar, S.F.; Nogueira, R.M.R.; Filippis, A.M.B.; et al. DENV-1 Genotype V in Brazil: Spatiotemporal dispersion pattern reveals continuous co-circulation of distinct lineages until 2016. Sci. Rep. 2018, 8, 17160. [Google Scholar] [CrossRef] [PubMed]
- San Martín, J.L.; Brathwaite, O.; Zambrano, B.; Solórzano, J.O.; Bouckenooghe, A.; Dayan, G.H.; Guzmán, M.G. The epidemiology of dengue in the Americas over the last three decades: A worrisome reality. Am. J. Trop. Med. Hyg. 2010, 82, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.A.M.; Pavon, J.A.R.; Dias, L.S.; Viniski, A.E.; Souza, C.L.C.; de Oliveira, E.C.; de Azevedo, V.C.; da Silva, S.P.; Cruz, A.C.R.; Medeiros, D.B.A.; et al. Dengue virus serotype 2 genotype III evolution during the 2019 outbreak in Mato Grosso, Midwestern Brazil. Infect. Genet. Evol. 2023, 113, 105487. [Google Scholar] [CrossRef] [PubMed]
- Yenamandra, S.P.; Koo, C.; Chiang, S.; Lim, H.S.J.; Yeo, Z.Y.; Ng, L.C.; Hapuarachchi, H.C. Evolution, heterogeneity and global spread of the cosmopolitan genotype of Dengue virus type 2. Sci. Rep. 2021, 11, 13496. [Google Scholar] [CrossRef]
- Torres, M.C.; Lima de Mendonça, M.C.; Damasceno Dos Santos Rodrigues, C.; Fonseca, V.; Ribeiro, M.S.; Brandão, A.P.; Venâncio da Cunha, R.; Dias, A.I.; Santos Vilas Boas, L.; Felix, A.C.; et al. Dengue Virus Serotype 2 Intrahost Diversity in Patients with Different Clinical Outcomes. Viruses 2021, 13, 349. [Google Scholar] [CrossRef]
- Vasconcelos, P.F.C.; Travassos da Rosa, E.S.; Travassos da Rosa, J.F.S.; Freitas, R.B.; Dégallier, N.; Rodrigues, S.G.; Travassos da Rosa, A.P.A. Epidemia de febre clássica de dengue causada pelo sorotipo 2 em Araguaiana, Tocantins, Brasil. Rev. Inst. Med. Trop. Sao Paulo 1993, 35, 141–148. [Google Scholar] [CrossRef]
- Souza, U.J.B.; Dos Santos, R.N.; Campos, F.S.; Lourenço, K.L.; Fonseca, F.G.; Spilki, F.R.; Corona-Ômica Br/McTi Network. High Rate of Mutational Events in SARS-CoV-2 Genomes across Brazilian Geographical Regions, February 2020 to June 2021. Viruses 2021, 13, 1806. [Google Scholar] [CrossRef]
- TOCANTINS. Secretaria de Saúde do Tocantins. Monitoramento das Arboviroses Urbanas (Dengue, Chikungunya e Zika) e Silvestre (Febre Amarela) no ano de 2020. Palmas—TO. Available online: https://central.to.gov.br/download/250434 (accessed on 6 October 2022).
- Brasil. Ministério da Saúde. Monitoramento dos Casos de Arboviroses Até a Semana Epidemiológica 52 de 2022. Available online: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/edicoes/2023/boletim-epidemiologico-volume-54-no-01/ (accessed on 6 February 2023).
- Brasil. Ministério da Saúde. Diretrizes Nacionais Para a Prevenção e Controle de Epidemias de Dengue. Brasilia—DF. Secretaria de Vigilância em Saúde. 2009. Available online: https://bvsms.saude.gov.br/bvs/publicacoes/diretrizes_nacionais_prevencao_controle_dengue.pdf (accessed on 5 December 2022).
- Brasil. Ministério da Saúde. Departamento de Vigilância Epidemiológica. Guia de Vigilância Epidemiológica. Brasilia—DF. Secretaria de Vigilância em Saúde. 2005. Available online: https://bvsms.saude.gov.br/bvs/publicacoes/Guia_Vig_Epid_novo2.pdf (accessed on 6 February 2023).
- Brazil. Ministry of Health of Brazil. Secretariat of Health Surveillance. Department of Noncommunicable Diseases Surveillance and Health Promotion. Health Brazil 2015/2016: An Analysis of Health Situation and the Epidemic Caused by Zika Virus and Other Diseases Transmitted by Aedes Aegypti, 2017. 132p. Available online: https://bvsms.saude.gov.br/bvs/publicacoes/health_brazil_2015_2016.pdf (accessed on 9 September 2023).
- Abrão, E.P.; Espósito, D.L.; Lauretti, F.; da Fonseca, B.A. Dengue vaccines: What we know, what has been done, but what does the future hold? Rev. Saude Publica 2015, 49, 60. [Google Scholar] [CrossRef]
- Gonçalves, C.M.; Melo, F.F.; Bezerra, J.M.; Chaves, B.A.; Silva, B.M.; Silva, L.D.; Pessanha, J.E.M.; Arias, J.R.; Secundino, N.F.C.; Norris, D.E.; et al. Distinct variation in vector competence among nine field populations of Aedes aegypti from a Brazilian dengue-endemic risk city. Parasites Vectors 2014, 7, 320. [Google Scholar] [CrossRef]
- Holmes, E.C.; Twiddy, S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003, 3, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Áñez, G. Evolución molecular del virus dengue: Un área de investigación prioritaria. Investig. Clínica 2007, 48, 273–276. Available online: http://ve.scielo.org/scielo.php?script=sci_arttext&pid=S0535-51332007000300001&lng=es (accessed on 6 February 2023).
- Santiago, G.A.; Vergne, E.; Quiles, Y.; Cosme, J.; Vazquez, J.; Medina, J.F.; Medina, F.; Colón, C.; Margolis, H.; Muñoz-Jordán, J.L. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl. Trop. Dis. 2013, 7, e2311. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, S.C.B.; Blacklaws, B.A.; Yohan, B.; Yudhaputri, F.A.; Hayati, R.F.; Schwem, B.; Salvaña, E.M.; Destura, R.V.; Lester, J.S.; Myint, K.S.; et al. Assessment of a multiplex PCR and Nanopore-based method for dengue virus sequencing in Indonesia. Virol. J. 2020, 17, 24. [Google Scholar] [CrossRef]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.M.; et al. Genome Detective: An automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree v1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 9 September 2023).
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using tempest (formerly path-o-gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekseyenko, A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Ho, S.Y.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 2013, 30, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Yu, G.; Lam, T.T.; Zhu, H.; Guan, Y. Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Fritsch, H.; Moreno, K.; Lima, I.A.B.; Santos, C.S.; Costa, B.G.G.; de Almeida, B.L.; Dos Santos, R.A.; Francisco, M.V.L.O.; Sampaio, M.P.S.; de Lima, M.M.; et al. Phylogenetic Reconstructions Reveal the Circulation of a Novel Dengue Virus-1V Clade and the Persistence of a Dengue Virus-2 III Genotype in Northeast Brazil. Viruses 2023, 15, 1073. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, A.; Manoharan, M. Chapter 16—Dengue Virus. Emerging and Reemerging Viral Pathogens; Academic Press: Cambridge, MA, USA, 2020; pp. 281–359. [Google Scholar] [CrossRef]
- TOCANTINS. Secretaria de Saúde do Tocantins. Monitoramento dos Casos de Dengue Até a Semana Epidemiológica 43. Palmas—TO. Boletins Epidemiológicos. 2022. Available online: https://central.to.gov.br/download/307814 (accessed on 3 January 2023).
- Salles, T.S.; da Encarnação Sá-Guimarães, T.; de Alvarenga, E.S.L.; Guimarães-Ribeiro, V.; de Meneses, M.D.F.; de Castro-Salles, P.F.; Dos Santos, C.R.; do Amaral Melo, A.C.; Soares, M.R.; Ferreira, D.F.; et al. History, epidemiology and diagnostics of dengue in the American and Brazilian contexts: A review. Parasit. Vectors 2018, 11, 264. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.B., Jr.; Martelli, C.M.; Coelho, G.E.; Simplicio, A.C.; Hatch, D.L. Dengue and dengue hemorrhagic fever, Brazil, 1981–2002. Emerg. Infect. Dis. 2005, 11, 48–53. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Scott, T.W.; Zou, C.; Pigott, D.M.; Duda, K.A.; Bhatt, S.; Katzelnick, L.; Howes, R.E.; Battle, K.E.; et al. Global spread of dengue virus types: Mapping the 70-year history. Trends Microbiol. 2014, 22, 138–146. [Google Scholar] [CrossRef]
- Araújo, H.R.; Carvalho, D.O.; Ioshino, R.S.; Costa-da-Silva, A.L.; Capurro, M.L. Aedes aegypti Control Strategies in Brazil: Incorporation of New Technologies to Overcome the Persistence of Dengue Epidemics. Insects 2015, 11, 576–594. [Google Scholar] [CrossRef]
- Santos, C.H.; Sousa, F.Y.; Lima, L.R.; Stival, M.M. Perfil Epidemiológico do Dengue em Anápolis-GO, 2001–2007. Rev. Pat. Trop. 2009, 38, 249–259. Available online: https://revistas.ufg.br/index.php/iptsp/article/view/8588/6071 (accessed on 6 March 2023). [CrossRef]
- Valadares, A.F.; Rodrigues, C.; Filho, J.; Peluzio, J.M. Impacto da dengue em duas principais cidades do Estado do Tocantins: Infestação e fator ambiental (2000 a 2010). Epidemiol. Serv. Saúde 2013, 22, 59–66. [Google Scholar] [CrossRef]
- Monteiro, E.S.C.; Coelho, M.E.; Cunha, I.S.; Cavalcante, M.A.S.; Carvalho, F.A.A. Aspectos epidemiológicos e vetoriais da dengue na cidade de Teresina, Piauí–Brasil, 2002 a 2006. Epidemiol. Serv. Saúde 2009, 18, 365–374. [Google Scholar] [CrossRef]
- Nogueira, R.M.; de Araújo, J.M.; Schatzmayr, H.G. Dengue viruses in Brazil, 1986-2006. Rev. Panam. Salud Publica 2007, 22, 358–363. [Google Scholar] [CrossRef]
- Chen, R.; Vasilakis, N. Dengue--quo tu et quo vadis? Viruses 2011, 3, 1562–1608. [Google Scholar] [CrossRef] [PubMed]
- Áñez, G.; Heisey, D.A.; Espina, L.M.; Stramer, S.L.; Rios, M. Phylogenetic analysis of dengue virus types 1 and 4 circulating in Puerto Rico and Key West, Florida, during 2010 epidemics. Am. J. Trop. Med. Hyg. 2012, 87, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, A.R.; Cruz, A.C.R.; Vallinoto, M.; Melo, D.V.; Ramos, R.T.J.; Medeiros, D.B.A.; Silva, E.V.P.; Vasconcelos, P.F.C. Molecular characterisation of dengue virus type 1 reveals lineage replacement during circulation in Brazilian territory. Mem. Inst. Oswaldo Cruz 2012, 107, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Drumond, B.P.; Mondini, A.; Schmidt, D.J.; Bosch, I.; Nogueira, M.L. Population dynamics of DENV-1 genotype V in Brazil is characterized by co-circulation and strain/lineage replacement. Arch. Virol. 2012, 157, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.E.; Martin, D.P.; Oliveira, L.M.; Ribeiro, B.M.; Nagata, T. Comparative analysis of American Dengue virus type 1 full-genome sequences. Virus Genes 2010, 40, 60–66. [Google Scholar] [CrossRef]
- Rico-Hesse, R.; Harrison, L.M.; Salas, R.A.; Tovar, D.; Nisalak, A.; Ramos, C.; Boshell, J.; de Mesa, M.T.; Nogueira, R.M.; da Rosa, A.T. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 1997, 230, 244–251. [Google Scholar] [CrossRef]
- Rico-Hesse, R. Dengue virus evolution and virulence models. Clin. Infect. Dis. 2007, 44, 1462–1466. [Google Scholar] [CrossRef]
- Torres, M.C.; de Bruycker Nogueira, F.; Fernandes, C.A.; Louzada Silva Meira, G.; Ferreira de Aguiar, S.; Chieppe, A.O.; Bispo de Filippis, A.M. Re-introduction of dengue virus serotype 2 in the state of Rio de Janeiro after almost a decade of epidemiological silence. PLoS ONE 2019, 14, e0225879. [Google Scholar] [CrossRef]
- Nogueira, R.M.; Miagostovich, M.P.; Lampe, E.; Schatzmayr, H.G. Isolation of dengue virus type 2 in Rio de Janeiro. Mem. Inst. Oswaldo Cruz 1990, 85, 253. [Google Scholar] [CrossRef]
- de Jesus, J.G.; Dutra, K.R.; Sales, F.C.D.S.; Claro, I.M.; Terzian, A.C.; Candido, D.D.S.; Hill, S.C.; Thézé, J.; Torres, C.; D’agostini, T.L.; et al. Genomic detection of a virus lineage replacement event of dengue virus serotype 2 in Brazil, 2019. Memórias Inst. Oswaldo Cruz 2020, 115, e190423. [Google Scholar] [CrossRef]
- Carrillo-Valenzo, E.; Danis-Lozano, R.; Velasco-Hernández, J.X.; Sánchez-Burgos, G.; Alpuche, C.; López, I.; Rosales, C.; Baronti, C.; de Lamballerie, X.; Holmes, E.C.; et al. Evolution of dengue virus in Mexico is characterized by frequent lineage replacement. Arch Virol. 2010, 155, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Mammen, M.P., Jr.; Chinnawirotpisan, P.; Klungthong, C.; Rodpradit, P.; Monkongdee, P.; Nimmannitya, S.; Kalayanarooj, S.; Holmes, E.C. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J. Virol. 2005, 79, 15123–15130. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Pereira, L.A.; Santiago, G.A.; Fonseca, V.; Mendoza, M.P.G.; de Oliveira, C.; de Moraes, L.; Xavier, J.; Tosta, S.; Fritsch, H.; et al. Emergence of Dengue Virus Serotype 2 Cosmopolitan Genotype, Brazil. Emerg. Infect. Dis. 2022, 28, 1725–1727. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.; Mayer, S.V.; Johnson, W.L.; Chen, R.; Volkova, E.; Vilcarromero, S.; Widen, S.G.; Wood, T.G.; Suarez-Ognio, L.; Long, K.C.; et al. Lineage II of Southeast Asian/American DENV-2 is associated with a severe dengue outbreak in the Peruvian Amazon. Am. J. Trop. Med. Hyg. 2014, 91, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Adelino, T.É.R.; Giovanetti, M.; Fonseca, V.; Xavier, J.; de Abreu, Á.S.; do Nascimento, V.A.; Demarchi, L.H.F.; Oliveira, M.A.A.; da Silva, V.L.; Cunha, G.M.; et al. Field and Classroom Initiatives for Portable Sequence-Based Monitoring of Dengue Virus in Brazil. Nat. Commun. 2021, 12, 2296. [Google Scholar] [CrossRef]
- Wispelaere, M.; Yang, P.L. Mutagenesis of the DI/DIII linker in dengue virus envelope protein impairs viral particle assembly. J. Virol. 2012, 86, 7072–7083. [Google Scholar] [CrossRef]
- Tay, M.Y.; Smith, K.; Ng, I.H.; Chan, K.W.; Zhao, Y.; Ooi, E.E.; Lescar, J.; Luo, D.; Jans, D.A.; Forwood, J.K.; et al. The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production. PLoS Pathog. 2012, 12, e1005886. [Google Scholar] [CrossRef]
- Rivera, L.; Biswal, S.; Sáez-Llorens, X.; Reynales, H.; López-Medina, E.; Borja-Tabora, C.; Bravo, L.; Sirivichayakul, C.; Kosalaraksa, P.; Martinez Vargas, L.; et al. Three-year Efficacy and Safety of Takeda’s Dengue Vaccine Candidate (TAK-003). Clin. Infect. Dis. 2022, 75, 107–117. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, U.J.B.d.; Macedo, Y.d.S.M.; Santos, R.N.d.; Cardoso, F.D.P.; Galvão, J.D.; Gabev, E.E.; Franco, A.C.; Roehe, P.M.; Spilki, F.R.; Campos, F.S. Circulation of Dengue Virus Serotype 1 Genotype V and Dengue Virus Serotype 2 Genotype III in Tocantins State, Northern Brazil, 2021–2022. Viruses 2023, 15, 2136. https://doi.org/10.3390/v15112136
Souza UJBd, Macedo YdSM, Santos RNd, Cardoso FDP, Galvão JD, Gabev EE, Franco AC, Roehe PM, Spilki FR, Campos FS. Circulation of Dengue Virus Serotype 1 Genotype V and Dengue Virus Serotype 2 Genotype III in Tocantins State, Northern Brazil, 2021–2022. Viruses. 2023; 15(11):2136. https://doi.org/10.3390/v15112136
Chicago/Turabian StyleSouza, Ueric José Borges de, Ygor da Silva Miranda Macedo, Raíssa Nunes dos Santos, Franciano Dias Pereira Cardoso, Jucimária Dantas Galvão, Evgeni Evgeniev Gabev, Ana Cláudia Franco, Paulo Michel Roehe, Fernando Rosado Spilki, and Fabrício Souza Campos. 2023. "Circulation of Dengue Virus Serotype 1 Genotype V and Dengue Virus Serotype 2 Genotype III in Tocantins State, Northern Brazil, 2021–2022" Viruses 15, no. 11: 2136. https://doi.org/10.3390/v15112136









