A Multiplex RT-PCR Method for the Detection of Reptarenavirus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Samples
2.2. Ethics Statement
2.3. Cell Culture and Viruses
2.4. Synthesis of Control RNAs
2.5. RNA Extraction
2.6. Quantitative Reverse Transcription PCR (qRT-PCR)
2.7. cDNA Synthesis and Multiplex RT-PCR
2.8. Immunocytochemistry
2.9. Statistical Analyses
3. Results
3.1. Primer Cocktail and Optimization of the Cycling Conditions for the Multiplex RT-PCR Method
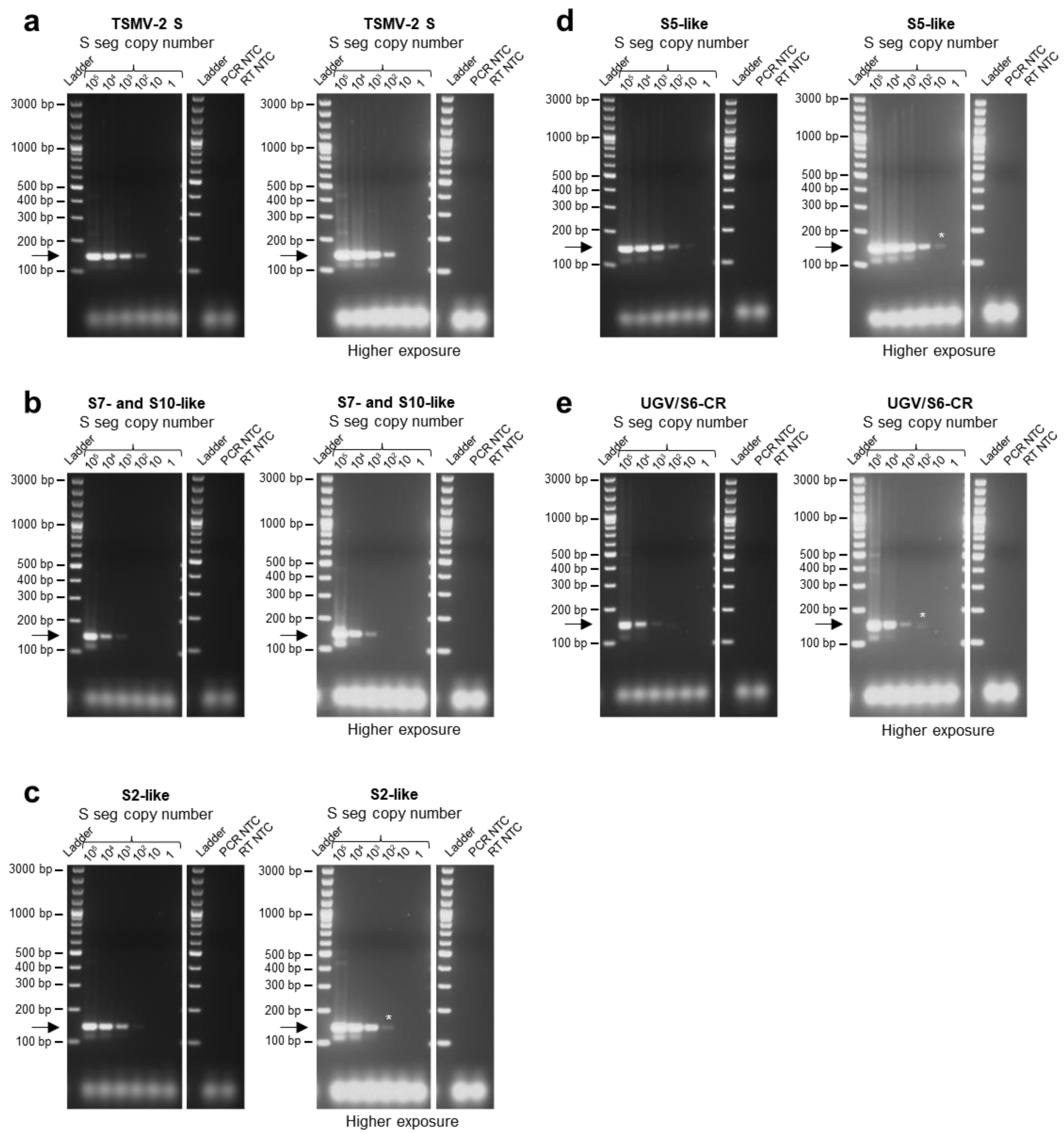
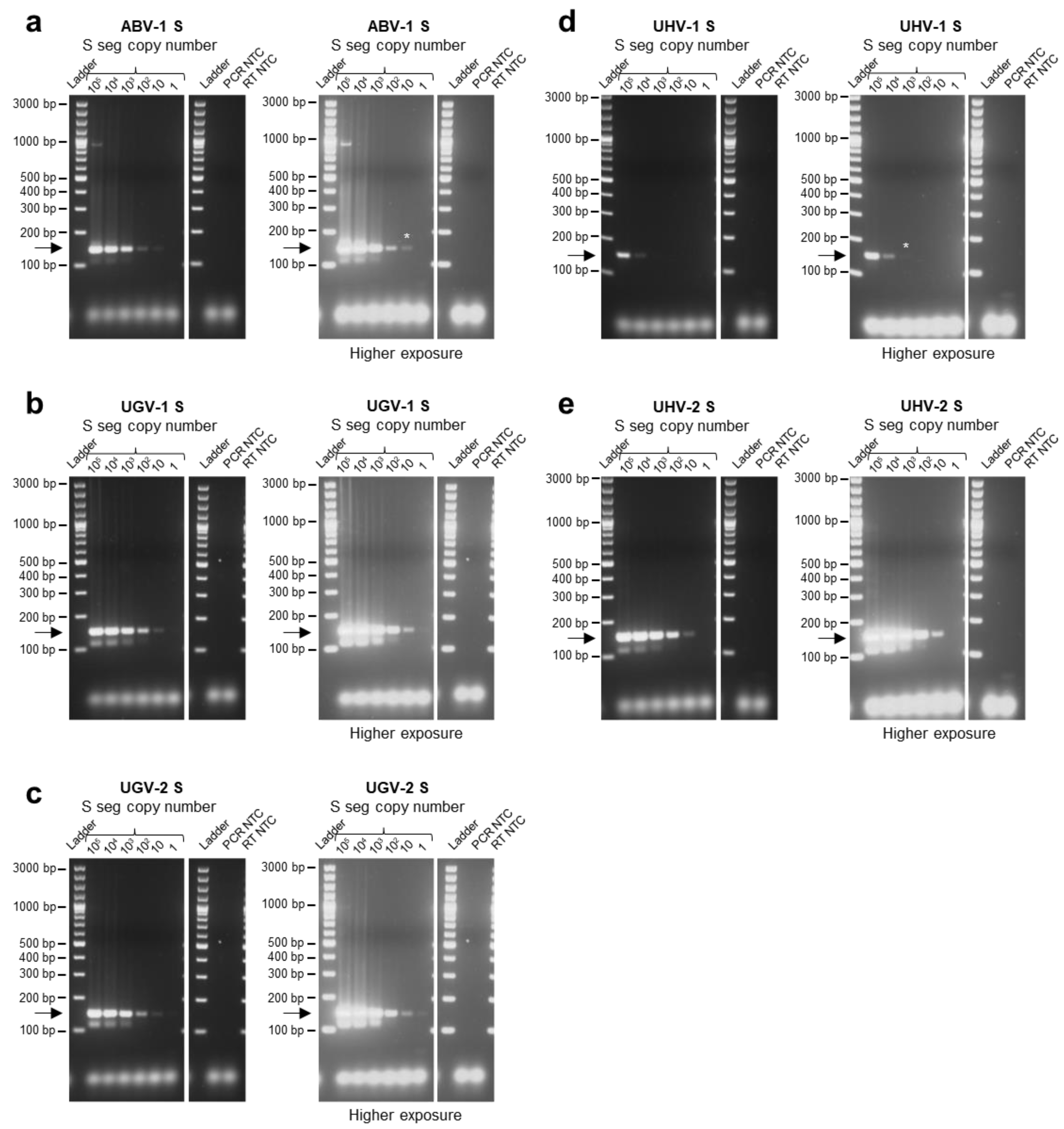
3.2. Limit of Detection of the Multiplex RT-PCR Protocol for Specific Reptarenavirus S Segments and Optimization of Primer Concentrations
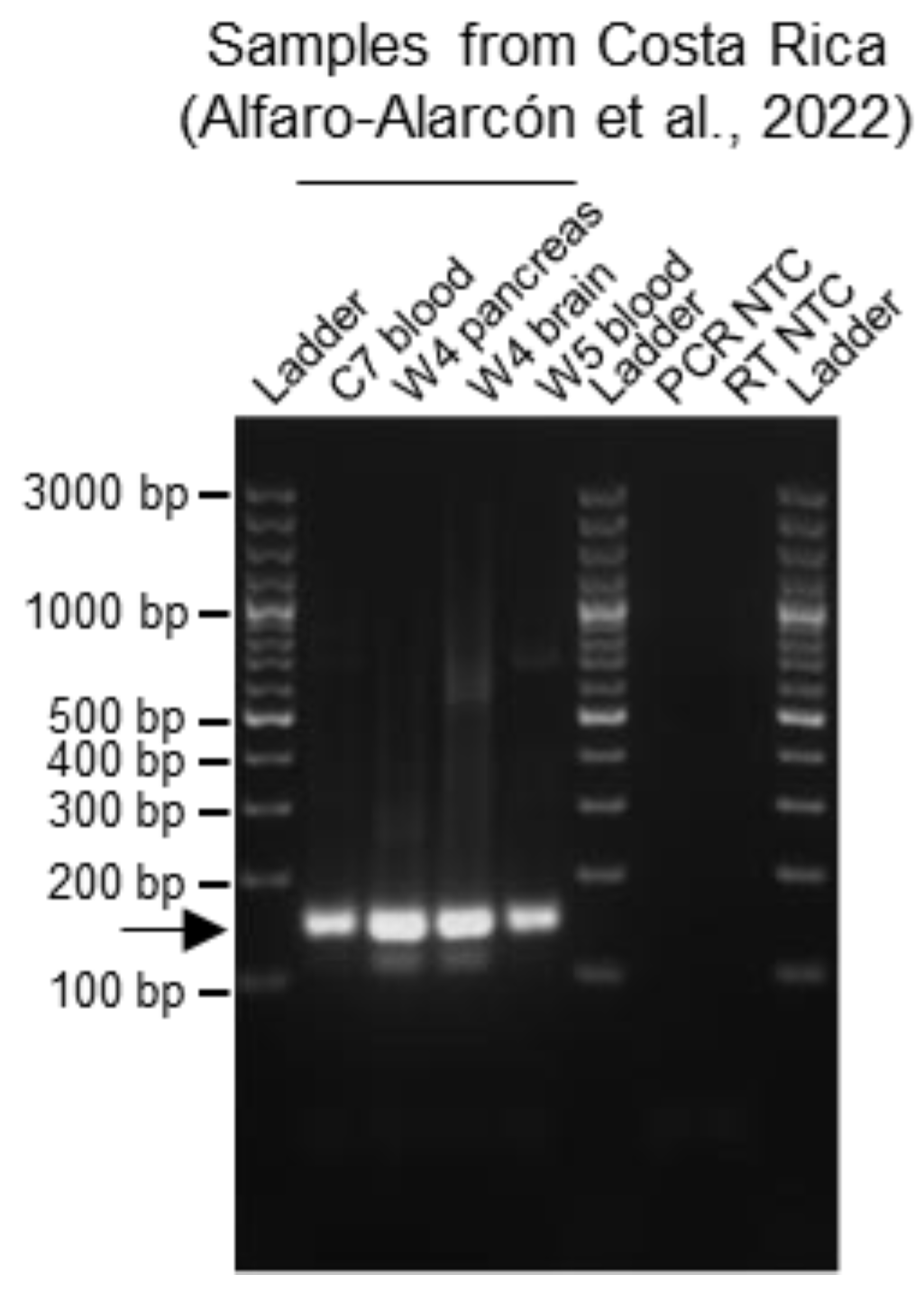
3.3. Validation of the Multiplex RT-PCR Protocol by Testing Its Reliability on Samples from Different Snake Collections
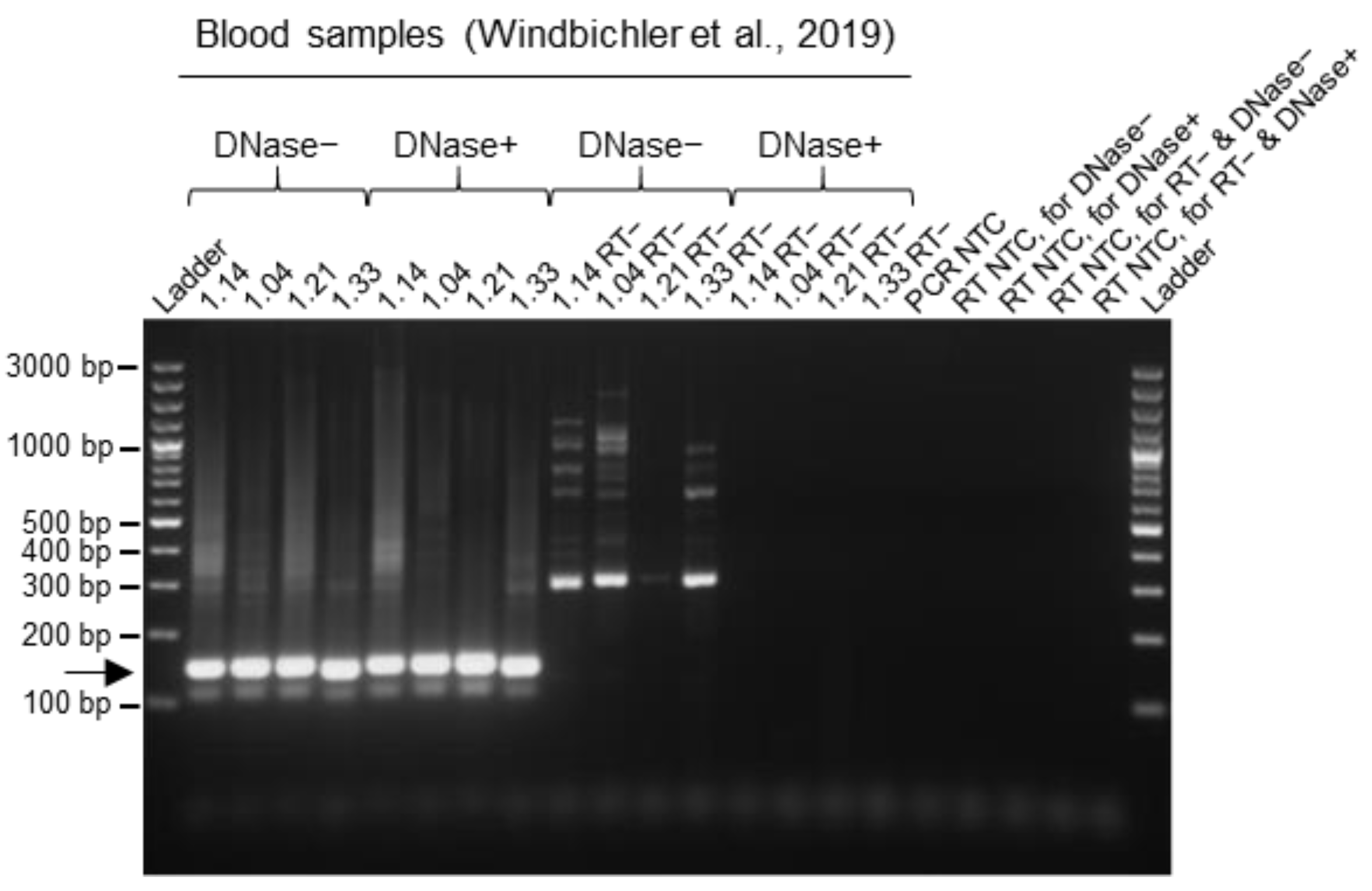
3.4. Improvement of Data Quality by DNase Treatment during the RNA Extraction Procedure from Blood Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schumacher, J.; Jacobson, E.R.; Homer, B.L.; Gaskin, J.M. Inclusion body disease in boid snakes. J. Zoo Wildl. Med. 1994, 25, 511–524. [Google Scholar]
- Chang, L.W.; Jacobson, E.R. Inclusion body disease, a worldwide infectious disease of boid snakes: A review. J. Exot. Pet Med. 2010, 19, 216–225. [Google Scholar] [CrossRef]
- Hetzel, U.; Sironen, T.; Laurinmäki, P.; Liljeroos, L.; Patjas, A.; Henttonen, H.; Vaheri, A.; Artelt, A.; Kipar, A.; Butcher, S.J.; et al. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J. Virol. 2013, 87, 10918–10935. [Google Scholar] [CrossRef] [PubMed]
- Aqrawi, T.; Stöhr, A.C.; Knauf-Witzens, T.; Krengel, A.; Heckers, K.O.; Marschang, R.E. Identification of snake arenaviruses in live boas and pythons in a zoo in Germany. Tierarztl. Prax. Ausg. K Kleintiere Heimtiere 2015, 43, 239–247. [Google Scholar] [CrossRef]
- Chang, L.; Fu, D.; Stenglein, M.D.; Hernandez, J.A.; DeRisi, J.L.; Jacobson, E.R. Detection and prevalence of boid inclusion body disease in collections of boas and pythons using immunological assays. Vet. J. 2016, 218, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Hetzel, U.; Sironen, T.; Korzyukov, Y.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Co-infecting reptarenaviruses can be vertically transmitted in Boa constrictor. PLoS Pathog. 2017, 13, e1006179. [Google Scholar] [CrossRef] [PubMed]
- Windbichler, K.; Michalopoulou, E.; Palamides, P.; Pesch, T.; Jelinek, C.; Vapalahti, O.; Kipar, A.; Hetzel, U.; Hepojoki, J. Antibody response in snakes with boid inclusion body disease. PLoS ONE 2019, 14, e0221863. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.; Marschang, R.E.; Leineweber, C.; Hellebuyck, T. Prevalence of inclusion body disease and associated comorbidity in captive collections of boid and pythonid snakes in Belgium. PLoS ONE 2020, 15, e0229667. [Google Scholar] [CrossRef] [PubMed]
- Thiele, T.; Baggio, F.; Prähauser, B.; Ruiz Subira, A.; Michalopoulou, E.; Kipar, A.; Hetzel, U.; Hepojoki, J. Reptarenavirus S segment RNA levels correlate with the presence of inclusion bodies and the number of L segments in snakes with reptarenavirus infection-Lessons learned from a large breeding colony. Microbiol. Spectr. 2023, 11, e0506522. [Google Scholar] [CrossRef]
- Alfaro-Alarcón, A.; Hetzel, U.; Smura, T.; Baggio, F.; Morales, J.A.; Kipar, A.; Hepojoki, J. Boid inclusion body disease is also a disease of wild boa constrictors. Microbiol. Spectr. 2022, 10, e0170522. [Google Scholar] [CrossRef]
- Wozniak, E.; McBride, J.; DeNardo, D.; Tarara, R.; Wong, V.; Osburn, B. Isolation and characterization of an antigenically distinct 68-kd protein from nonviral intracytoplasmic inclusions in Boa constrictors chronically infected with the inclusion body disease virus (IBDV: Retroviridae). Vet. Pathol. 2000, 37, 449–459. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Sanders, C.; Kistler, A.L.; Ruby, J.G.; Franco, J.Y.; Reavill, D.R.; Dunker, F.; Derisi, J.L. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: Candidate etiological agents for snake inclusion body disease. mBio 2012, 3, e00180-12. [Google Scholar] [CrossRef]
- Bodewes, R.; Kik, M.J.L.; Raj, V.S.; Schapendonk, C.M.E.; Haagmans, B.L.; Smits, S.L.; Osterhaus, A.D.M.E. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J. Gen. Virol. 2013, 94 Pt 6, 1206–1210. [Google Scholar] [CrossRef]
- Radoshitzky, S.R.; Bào, Y.; Buchmeier, M.J.; Charrel, R.N.; Clawson, A.N.; Clegg, C.S.; DeRisi, J.L.; Emonet, S.; Gonzalez, J.P.; Kuhn, H.; et al. Past, present, and future of arenavirus taxonomy. Arch. Virol. 2015, 160, 1851–1874. [Google Scholar] [CrossRef]
- Radoshitzky, S.R.; Buchmeier, M.J.; Charrel, R.N.; Gonzalez, J.P.; Günther, S.; Hepojoki, J.; Kuhn, J.H.; Lukashevich, I.S.; Romanowski, V.; Salvato, M.S.; et al. ICTV virus taxonomy profile: Arenaviridae. J. Gen. Virol. 2023, 104, 001891. [Google Scholar] [CrossRef]
- Chang, L.W.; Fu, A.; Wozniak, E.; Chow, M.; Duke, D.G.; Green, L.; Kelley, K.; Hernandez, J.A.; Jacobson, E.R. Immunohistochemical detection of a unique protein within cells of snakes having inclusion body disease, a world-wide disease seen in members of the families Boidae and Pythonidae. PLoS ONE 2013, 8, e82916. [Google Scholar] [CrossRef]
- Keilwerth, M.; Bühler, I.; Hoffmann, R.; Soliman, H.; El-Matbouli, M. Einschlusskörperchenkrankheit der Riesenschlangen (Inclusion Body Disease, IBD)-Eine hämatologische, histologische und elektronenmikroskopische Studie zur Diagnosefindung [Inclusion Body Disease (IBD of Boids)-A haematological, histological and electron microscopical study]. Berl. Munch. Tierarztl. Wochenschr. 2012, 125, 411–417. [Google Scholar]
- Argenta, F.F.; Hepojoki, J.; Smura, T.; Szirovicza, L.; Hammerschmitt, M.E.; Driemeier, D.; Kipar, A.; Hetzel, U. Identification of reptarenaviruses, hartmaniviruses, and a novel chuvirus in captive native brazilian boa constrictors with boid inclusion body disease. J. Virol. 2020, 94, e00001-20. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Sanchez-Migallon Guzman, D.; Garcia, V.E.; Layton, M.L.; Hoon-Hanks, L.L.; Boback, S.M.; Keel, M.K.; Drazenovich, T.; Hawkins, M.G.; DeRisi, J.L. Differential disease susceptibilities in experimentally reptarenavirus-infected boa constrictors and ball pythons. J. Virol. 2017, 91, e00451-17. [Google Scholar] [CrossRef]
- Hetzel, U.; Korzyukov, Y.; Keller, S.; Szirovicza, L.; Pesch, T.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Experimental reptarenavirus infection of Boa constrictor and Python regius. J. Virol. 2021, 95, e01968-20. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Jacobson, E.R.; Chang, L.W.; Sanders, C.; Hawkins, M.G.; Guzman, D.S.; Drazenovich, T.; Dunker, F.; Kamaka, E.K.; Fisher, D.; et al. Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog. 2015, 11, e1004900. [Google Scholar] [CrossRef] [PubMed]
- Hepojoki, J.; Salmenperä, P.; Sironen, T.; Hetzel, U.; Korzyukov, Y.; Kipar, A.; Vapalahti, O. Arenavirus coinfections are common in snakes with boid inclusion body disease. J. Virol. 2015, 89, 8657–8660. [Google Scholar] [CrossRef]
- Pontremoli, C.; Forni, D.; Cagliani, R.; Sironi, M. Analysis of Reptarenavirus genomes indicates different selective forces acting on the S and L segments and recent expansion of common genotypes. Infect. Genet. Evol. 2018, 64, 212–218. [Google Scholar] [CrossRef]
- Baggio, F.; Hetzel, U.; Nufer, L.; Kipar, A.; Hepojoki, J. A subpopulation of arenavirus nucleoprotein localizes to mitochondria. Sci. Rep. 2021, 11, 21048. [Google Scholar] [CrossRef]
- Lintala, A.; Szirovicza, L.; Kipar, A.; Hetzel, U.; Hepojoki, J. Persistent reptarenavirus and hartmanivirus infection in cultured boid cells. Microbiol. Spectr. 2022, 10, e0158522. [Google Scholar] [CrossRef]
- Hepojoki, J.; Hepojoki, S.; Smura, T.; Szirovicza, L.; Dervas, E.; Prähauser, B.; Nufer, L.; Schraner, E.M.; Vapalahti, O.; Kipar, A.; et al. Characterization of Haartman Institute snake virus-1 (HISV-1) and HISV-like viruses-The representatives of genus Hartmanivirus, family Arenaviridae. PLoS Pathog. 2018, 14, e1007415. [Google Scholar] [CrossRef]
- Hepojoki, J.; Kipar, A.; Korzyukov, Y.; Bell-Sakyi, L.; Vapalahti, O.; Hetzel, U. Replication of boid inclusion body disease-associated arenaviruses is temperature sensitive in both boid and mammalian cells. J. Virol. 2015, 89, 1119–1128. [Google Scholar] [CrossRef]
- Cheung, A.; Dufour, S.; Jones, G.; Kostoulas, P.; Stevenson, M.A.; Singanallur, N.B.; Firestone, S.M. Bayesian latent class analysis when the reference test is imperfect. Rev. Sci. Tech. 2021, 40, 271–286. [Google Scholar] [CrossRef]

| Primers | Reptarenavirus S Segment Sequences Expected to be Bound by the Primers | |||||||
|---|---|---|---|---|---|---|---|---|
| Name | Sequence | Length (nt) | Final Concentration (µM) | Tm | 100% Identity | 1 Mismatch | 2 Mismatches | 3 Mismatches |
| F1 | 5′-GTGCAGGCATAACCAATTCAC-3′ | 21 | 0.4 | 62.8 | UGV-1-4/S6, S6A, S7, S8, S9, S10 | ABV-2, other UGV-1-4/S6, S6B, S7-like, other S9, S10-like, UGV/S6-CR-wild4 | UHV-2, other UGV-1-4/S6, S2, S2-like, S4, GGV, NL-3 | UHV-3, PAV-1 |
| F2 | 5′-GTGCAGGCATAACAAATTCAC-3′ | 21 | 0.4 | 61.0 | S7-like, S10-like | UGV-1-4/S6, S6A, GGV, S2, S2-like, S4, S7, S8, S9, S10 | PAV-1, UHV-2, other UGV-1-4/S6, S6B, ABV-2, NL-3, other S9, UGV/S6-CR | Other UGV-1-4/S6 |
| F3 | 5′-CAGTGCTGGAATAACTAATTCTC-3′ | 23 | 0.4 | 59.8 | S11 | |||
| F4 | 5′-GTGCAGGCATAACCAACTC-3′ | 19 | 0.4 | 61.5 | TSMV-2, ArBV-1, ABV-2, UGV-1-4/S6, S9, UGV/S6-CR-wild4 | Other ArBV-1, S2, S2-like, S3, S4, GGV, NL-3, ABV-1, other UGV-1-4/S6 | PAV-1, UHV-1 | |
| F5 | 5′-GTGTTGGCATCACAAACTC-3′ | 19 | 0.4 | 59.8 | S5-like, S5 | PAV-1 | S2, S2-like, S4, GGV | |
| F6 | 5′-GCGCAGGCATTACCAATTC-3′ | 19 | 0.2 | 61.4 | UGV/S6-CR | UGV-1-4/S6, S6A, S7, S8, S9, S10 | Other UGV1-4/S6, S7-like, S10-like, UHV-2 | |
| R1 | 5′-CATGATCTCTAATTTCTCCTTCTTC-3′ | 25 | 0.4 | 59.9 | UGV-1-4/S6 | ABV-2, other UGV-1-4/S6, S6A, S7, S8, ArBV-1, UGV/S6-CR-wild4 | Other ABV-2, TSMV-2, PAV-1, GGV, other ArBV-1, S2, S3 | ABV-1, NL-3, other S2, S2-like, S4 |
| R2 | 5′-GATCTCTGATTTCACCTTCTTCC-3′ | 23 | 0.4 | 61.4 | PAV-1, ABV-2, TSMV-2, GGV, S2 | other S2, S2-like, ABV-1, other ABV-2, NL-3, S4 | ||
| R3 | 5′-TCTCTGATCTCACCTTCCTC-3′ | 20 | 0.4 | 60.6 | S6B, S9, S10 | S11, ArBV-1 | Other ArBV-1, S5, S5-like, UGV/S6-CR, UGV-1-4/S6 | UHV-2, UHV-3 |
| R4 | 5′-ACATGATCTCTAATCTCTCCTTC-3′ | 23 | 0.4 | 59.7 | UGV-1-4/S6, S6A, S7, S8, UGV/S6-CR-wild4 | Other UGV-1-4/S6, S7-like, S10-like | Other UGV-1-4/S6, ABV-1, ABV-2, ArBV-1, S2, S2-like, S4, S11 | Other ABV-2, other ArBV-1, GGV, PAV-1, other S2, S3, TMSV-2 |
| R5 | 5′-ATGACATGATCCCTAATCTCAC-3′ | 22 | 0.4 | 60.1 | UHV-2 | UHV-3 | UHV-1, ABV-1, S11 | ArBV-1, ABV-2, GGV |
| R6 | 5′-GGTCTCTTATTTCACCTTCCTC-3′ | 22 | 0.4 | 60.8 | S5-like, S5 | ArBV-1, S6B, S9, S10, S11 | S7-like, S10-like | |
| R7 | 5′-ATGACATGATCTCTGATCTCG-3′ | 21 | 0.2 | 58.7 | UGV/S6-CR | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baggio, F.; Hetzel, U.; Prähauser, B.; Dervas, E.; Michalopoulou, E.; Thiele, T.; Kipar, A.; Hepojoki, J. A Multiplex RT-PCR Method for the Detection of Reptarenavirus Infection. Viruses 2023, 15, 2313. https://doi.org/10.3390/v15122313
Baggio F, Hetzel U, Prähauser B, Dervas E, Michalopoulou E, Thiele T, Kipar A, Hepojoki J. A Multiplex RT-PCR Method for the Detection of Reptarenavirus Infection. Viruses. 2023; 15(12):2313. https://doi.org/10.3390/v15122313
Chicago/Turabian StyleBaggio, Francesca, Udo Hetzel, Barbara Prähauser, Eva Dervas, Eleni Michalopoulou, Tanja Thiele, Anja Kipar, and Jussi Hepojoki. 2023. "A Multiplex RT-PCR Method for the Detection of Reptarenavirus Infection" Viruses 15, no. 12: 2313. https://doi.org/10.3390/v15122313






