Emerging Prognostic and Predictive Significance of Stress Keratin 17 in HPV-Associated and Non HPV-Associated Human Cancers: A Scoping Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Method Selection
2.2. Eligibility
2.3. Information Sources and Searches
2.4. Study Selection
2.5. Data Collection and Synthesis of Results
2.6. Quality Assessment and Risk of Bias
3. Results
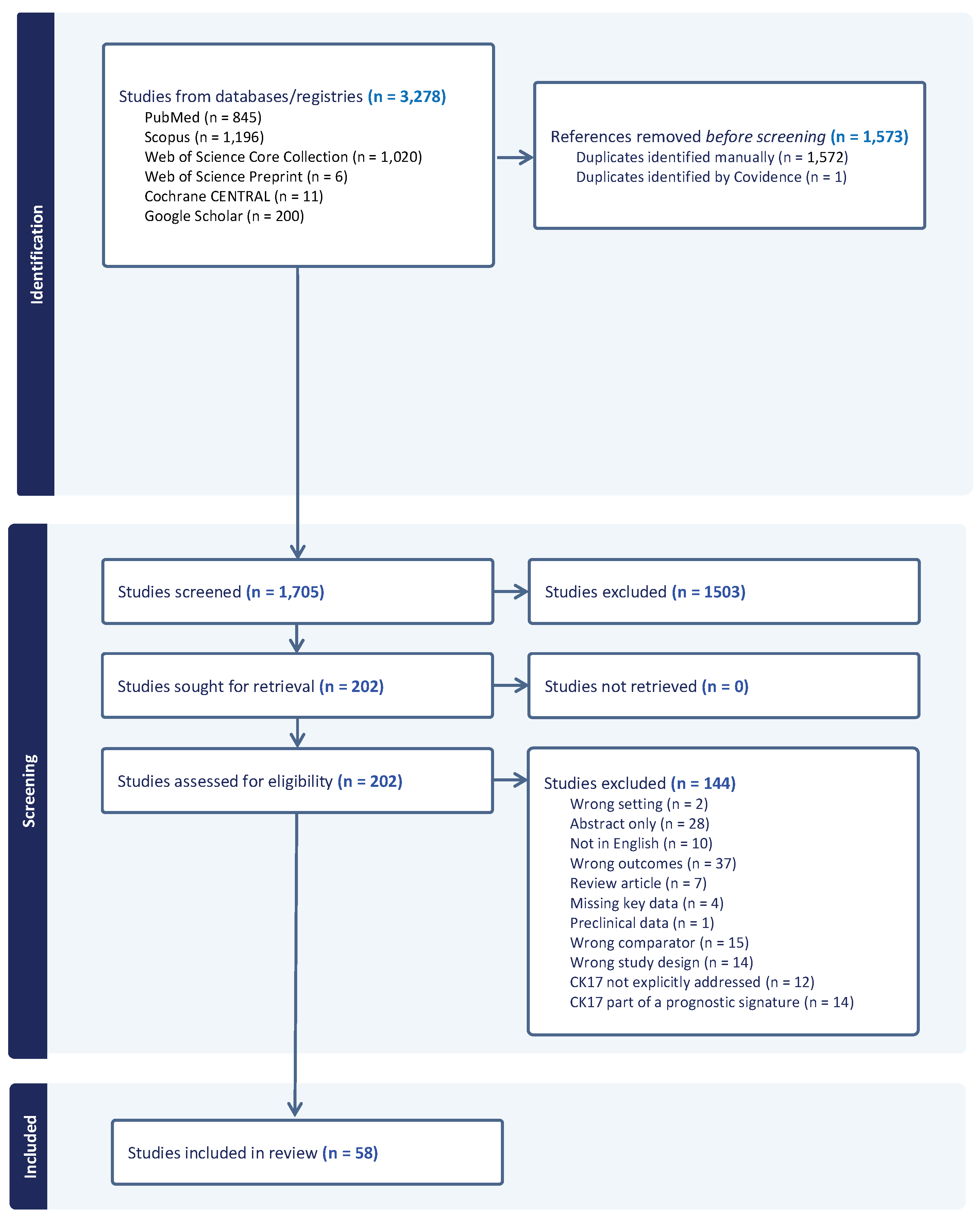
3.1. Literature Search
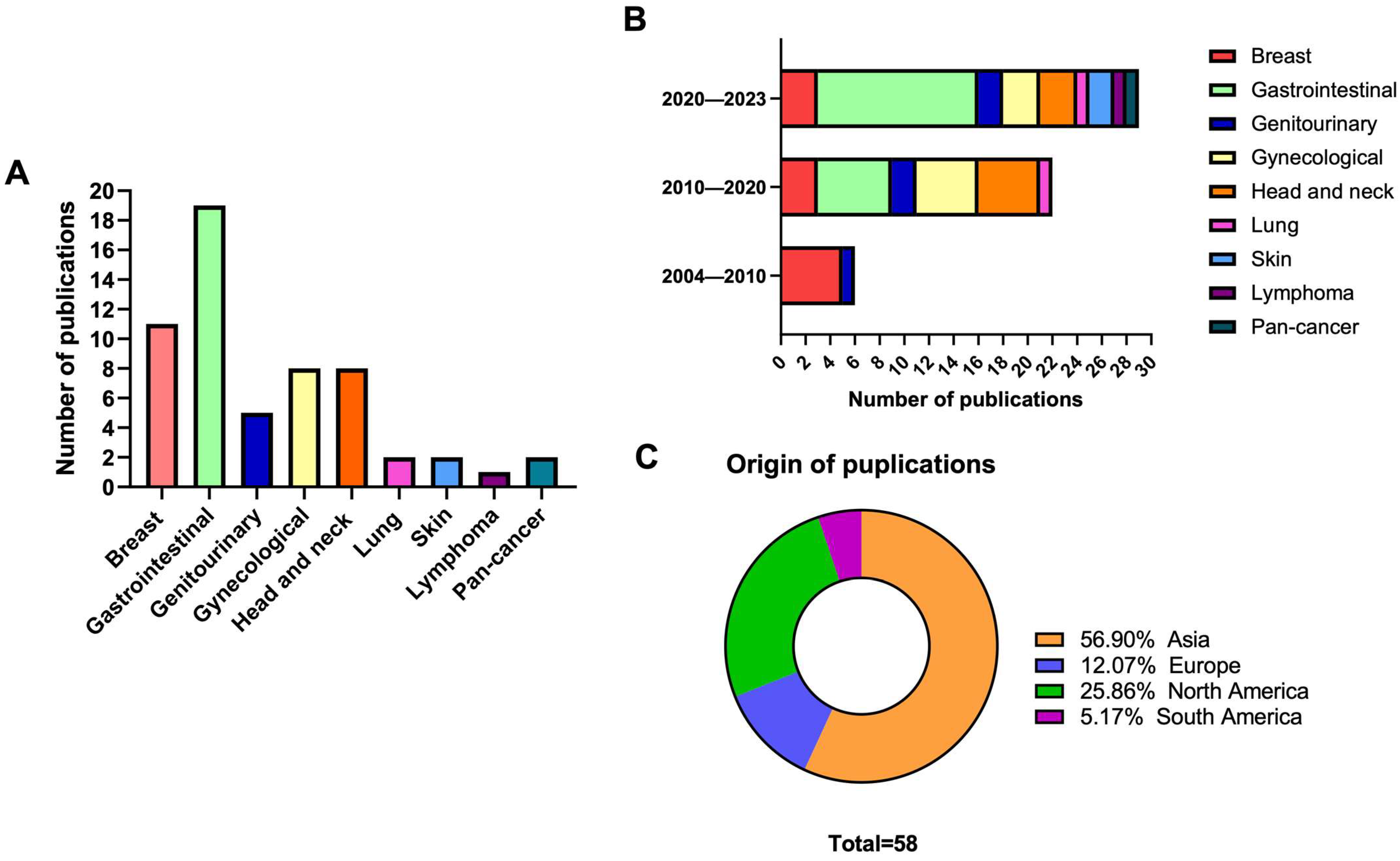
3.2. Included Studies’ Characteristics
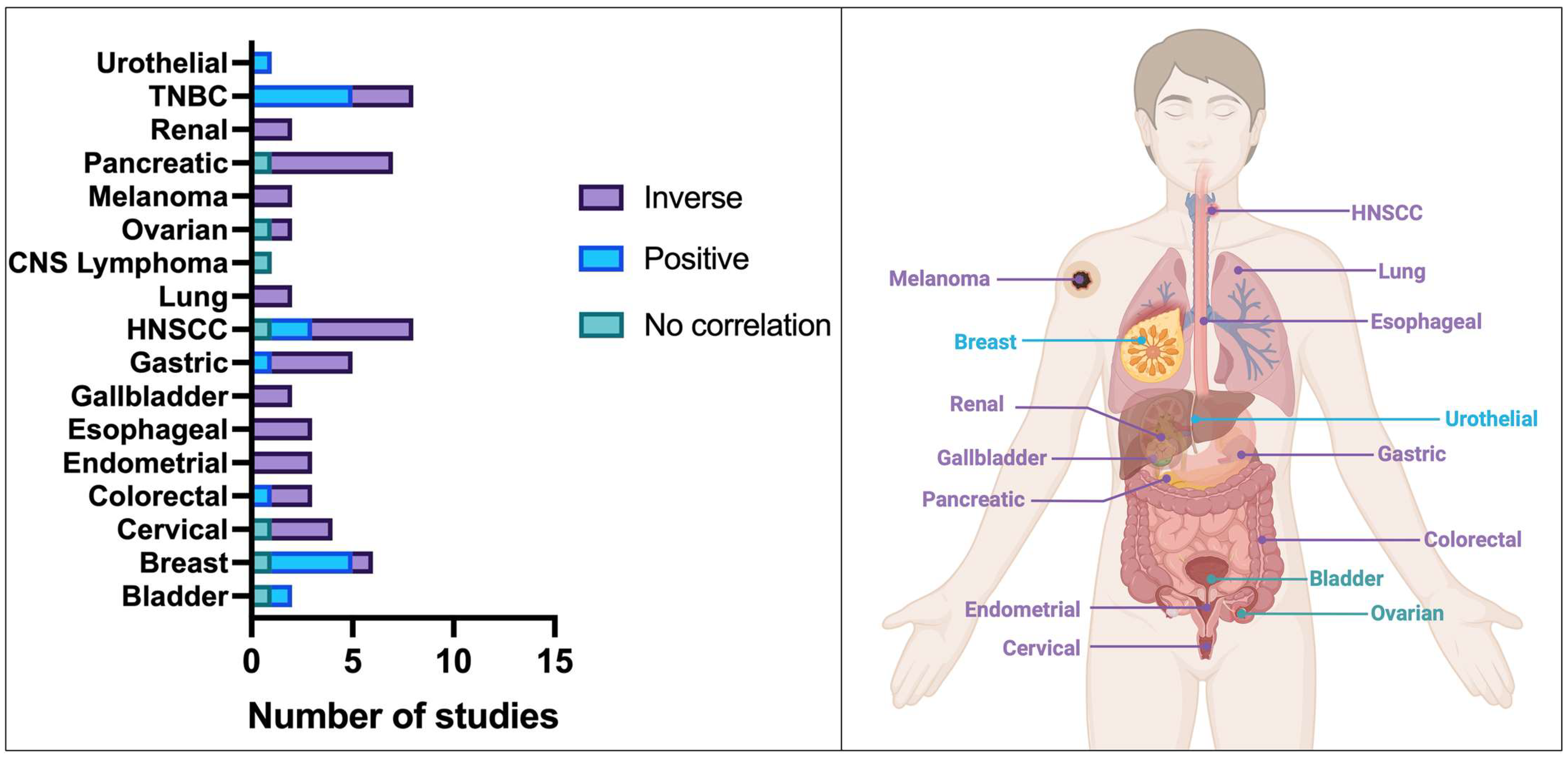
3.2.1. Prognostic Significance
3.2.2. Predictive Significance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Fountzilas, E.; Nikanjam, M.; Kurzrock, R. Review of Precision Cancer Medicine: Evolution of the Treatment Paradigm. Cancer Treat. Rev. 2020, 86, 102019. [Google Scholar] [CrossRef]
- Ballman, K.V. Biomarker: Predictive or Prognostic? J. Clin. Oncol. 2015, 33, 3968–3971. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Baraks, G.; Tseng, R.; Pan, C.H.; Kasliwal, S.; Leiton, C.V.; Shroyer, K.R.; Escobar-Hoyos, L.F. Dissecting the Oncogenic Roles of Keratin 17 in the Hallmarks of Cancer. Cancer Res. 2022, 82, 1159–1166. [Google Scholar] [CrossRef]
- Chu, P.G.; Weiss, L.M. Keratin Expression in Human Tissues and Neoplasms. Histopathology 2002, 40, 403–439. [Google Scholar] [CrossRef]
- Moll, R.; Franke, W.W.; Schiller, D.L.; Geiger, B.; Krepler, R. The Catalog of Human Cytokeratins: Patterns of Expression in Normal Epithelia, Tumors and Cultured Cells. Cell 1982, 31, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Babu, S.; Mockler, D.C.; Roa-Peña, L.; Szygalowicz, A.; Kim, N.W.; Jahanfard, S.; Gholami, S.S.; Moffitt, R.; Fitzgerald, J.P.; Escobar-Hoyos, L.F.; et al. Keratin 17 Is a Sensitive and Specific Biomarker of Urothelial Neoplasia. Mod. Pathol. 2019, 32, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Hoyos, L.F.; Shah, R.; Roa-Peña, L.; Vanner, E.A.; Najafian, N.; Banach, A.; Nielsen, E.; Al-Khalil, R.; Akalin, A.; Talmage, D.; et al. Keratin-17 Promotes p27KIP1 Nuclear Export and Degradation and Offers Potential Prognostic Utility. Cancer Res. 2015, 75, 3650–3662. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ni, X.F.; Tang, H.; Zhang, J.; Zheng, C.; Lin, J.; Wang, C.; Sun, L.; Chen, B. Krt17 Functions as a Tumor Promoter and Regulates Proliferation, Migration and Invasion in Pancreatic Cancer via Mtor/S6k1 Pathway. Cancer Manag. Res. 2020, 12, 2087–2095. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Yang, C.; Hu, W.; Chen, T.; Wang, Q.; Pan, F.; Qiu, B.; Tang, B. Knockdown of KRT17 Decreases Osteosarcoma Cell Proliferation and the Warburg Effect via the AKT/mTOR/HIF1α Pathway. Oncol. Rep. 2020, 44, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Jang, T.H.; Huang, W.C.; Tung, S.L.; Lin, S.C.; Chen, P.M.; Cho, C.Y.; Yang, Y.Y.; Yen, T.C.; Lo, G.H.; Chuang, S.E.; et al. MicroRNA-485-5p Targets Keratin 17 to Regulate Oral Cancer Stemness and Chemoresistance via the Integrin/FAK/Src/ERK/β-Catenin Pathway. J. Biomed. Sci. 2022, 29, 42. [Google Scholar] [CrossRef]
- Wang, W.; Uberoi, A.; Spurgeon, M.; Gronski, E.; Majerciak, V.; Lobanov, A.; Hayes, M.; Loke, A.; Zheng, Z.M.; Lambert, P.F. Stress Keratin 17 Enhances Papillomavirus Infection-Induced Disease by Downregulating T Cell Recruitment. PLoS Pathog. 2020, 16, e1008206. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lozar, T.; Golfinos, A.E.; Lee, D.; Gronski, E.; Ward-Shaw, E.; Hayes, M.; Bruce, J.Y.; Kimple, R.J.; Hu, R.; et al. Stress Keratin 17 Expression in Head and Neck Cancer Contributes to Immune Evasion and Resistance to Immune-Checkpoint Blockade. Clin. Cancer Res. 2022, 28, 2953–2968. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, S.; Wang, G. Keratin 17 in Disease Pathogenesis: From Cancer to Dermatoses. J. Pathol. 2019, 247, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Spurgeon, M.E.; Pope, A.; McGregor, S.; Ward-Shaw, E.; Gronski, E.; Lambert, P.F. Stress Keratin 17 and Estrogen Support Viral Persistence and Modulate the Immune Environment during Cervicovaginal Murine Papillomavirus Infection. Proc. Natl. Acad. Sci. USA 2023, 120, e2214225120. [Google Scholar] [CrossRef]
- Nazarian, R.M.; Primiani, A.; Doyle, L.A.; Linskey, K.R.; Duncan, L.M.; Odze, R.D.; Zukerberg, L.R. Cytokeratin 17: An Adjunctive Marker of Invasion in Squamous Neoplastic Lesions of the Anus. Am. J. Surg. Pathol. 2014, 38, 78–85. [Google Scholar] [CrossRef]
- Lo, B.K.K.; Yu, M.; Zloty, D.; Cowan, B.; Shapiro, J.; McElwee, K.J. CXCR3/Ligands Are Significantly Involved in the Tumorigenesis of Basal Cell Carcinomas. Am. J. Pathol. 2010, 176, 2435–2446. [Google Scholar] [CrossRef]
- Hobbs, R.P.; Batazzi, A.S.; Han, M.C.; Coulombe, P.A. Loss of Keratin 17 Induces Tissue-Specific Cytokine Polarization and Cellular Differentiation in HPV16-Driven Cervical Tumorigenesis in Vivo. Oncogene 2016, 35, 5653–5662. [Google Scholar] [CrossRef]
- Liu, L.; Sun, L.; Zheng, J.; Cui, L. Berberine Modulates Keratin 17 to Inhibit Cervical Cancer Cell Viability and Metastasis. J. Recept. Signal Transduct. 2020, 41, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Hoyos, L.F.; Yang, J.; Zhu, J.; Cavallo, J.A.; Zhai, H.; Burke, S.; Koller, A.; Chen, E.I.; Shroyer, K.R. Keratin 17 in Premalignant and Malignant Squamous Lesions of the Cervix: Proteomic Discovery and Immunohistochemical Validation as a Diagnostic and Prognostic Biomarker. Mod. Pathol. 2014, 27, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Mockler, D.; Escobar-Hoyos, L.F.; Akalin, A.; Romeiser, J.; Laurie Shroyer, A.; Shroyer, K.R. Keratin 17 Is a Prognostic Biomarker in Endocervical Glandular Neoplasia. Am. J. Clin. Pathol. 2017, 148, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Regenbogen, E.; Mo, M.; Romeiser, J.; Shroyer, A.L.W.; Escobar-Hoyos, L.F.; Burke, S.; Shroyer, K.R. Elevated Expression of Keratin 17 in Oropharyngeal Squamous Cell Carcinoma Is Associated with Decreased Survival. Head Neck 2018, 40, 1788–1798. [Google Scholar] [CrossRef]
- He, X.; Marchionni, L.; Hansel, D.E.; Yu, W.; Sood, A.; Yang, J.; Parmigiani, G.; Matsui, W.; Berman, D.M. Differentiation of a Highly Tumorigenic Basal Cell Compartment in Urothelial Carcinoma. Stem Cells 2009, 27, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Ide, M.; Kato, T.; Ogata, K.; Mochiki, E.; Kuwano, H.; Oyama, T. Keratin 17 Expression Correlates with Tumor Progression and Poor Prognosis in Gastric Adenocarcinoma. Ann. Surg. Oncol. 2012, 19, 3506–3514. [Google Scholar] [CrossRef]
- Wang, Y.F.; Lang, H.Y.; Yuan, J.; Wang, J.; Wang, R.; Zhang, X.H.; Zhang, J.; Zhao, T.; Li, Y.R.; Liu, J.Y.; et al. Overexpression of Keratin 17 Is Associated with Poor Prognosis in Epithelial Ovarian Cancer. Tumor Biol. 2013, 39, 1685–1689. [Google Scholar] [CrossRef]
- Thike, A.A.; Cheok, P.Y.; Jara-Lazaro, A.R.; Tan, B.; Tan, P.; Tan, P.H. Triple-Negative Breast Cancer: Clinicopathological Characteristics and Relationship with Basal-like Breast Cancer. Mod. Pathol. 2010, 23, 123–133. [Google Scholar] [CrossRef]
- Merkin, R.D.; Vanner, E.A.; Romeiser, J.L.; Shroyer, A.L.W.; Escobar-Hoyos, L.F.; Li, J.; Powers, R.S.; Burke, S.; Shroyer, K.R. Keratin 17 Is Overexpressed and Predicts Poor Survival in Estrogen Receptor–Negative/Human Epidermal Growth Factor Receptor-2–Negative Breast Cancer. Hum. Pathol. 2017, 62, 23–32. [Google Scholar] [CrossRef]
- Bai, J.D.K.; Babu, S.; Roa-Peña, L.; Hou, W.; Akalin, A.; Escobar-Hoyos, L.F.; Shroyer, K.R. Keratin 17 Is a Negative Prognostic Biomarker in High-Grade Endometrial Carcinomas. Hum. Pathol. 2019, 94, 40–50. [Google Scholar] [CrossRef]
- Roa-Peña, L.; Leiton, C.V.; Babu, S.; Pan, C.H.; Vanner, E.A.; Akalin, A.; Bandovic, J.; Moffitt, R.A.; Shroyer, K.R.; Escobar-Hoyos, L.F. Keratin 17 Identifies the Most Lethal Molecular Subtype of Pancreatic Cancer. Sci. Rep. 2019, 9, 11239. [Google Scholar] [CrossRef]
- Methodology for JBI Scoping Reviews. Available online: https://www.researchgate.net/publication/294736492_Methodology_for_JBI_Scoping_Reviews (accessed on 15 January 2023).
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Wang, Y.; Ping, J.; Xu, W.; Fang, W.; Liu, J. The Serum CK17 and CK19 Expressions in Cervical Cancer Patients and Their Prognostic Value. Am. J. Transl. Res. 2021, 13, 6439–6445. [Google Scholar] [PubMed]
- Roa-Peña, L.; Babu, S.; Leiton, C.V.; Wu, M.; Taboada, S.; Akalin, A.; Buscaglia, J.; Escobar-Hoyos, L.F.; Shroyer, K.R. Keratin 17 Testing in Pancreatic Cancer Needle Aspiration Biopsies Predicts Survival. Cancer Cytopathol. 2021, 129, 865–873. [Google Scholar] [CrossRef]
- Langner, C.; Wegscheider, B.J.; Rehak, P.; Ratschek, M.; Zigeuner, R. Prognostic Value of Keratin Subtyping in Transitional Cell Carcinoma of the Upper Urinary Tract. Virchows Arch. 2004, 445, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pinilla, S.M.; Jones, R.L.; Lambros, M.B.K.; Arriola, E.; Savage, K.; James, M.; Pinder, S.E.; Reis-Filho, J.S. MYC Amplification in Breast Cancer: A Chromogenic in Situ Hybridisation Study. J. Clin. Pathol. 2007, 60, 1017. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, R.; Toyoshima, T.; Tanaka, H.; Kawano, S.; Matsubara, R.; Goto, Y.; Jinno, T.; Maruse, Y.; Oobu, K.; Nakamura, S. Cytokeratin 17 mRNA as a Prognostic Marker of Oral Squamous Cell Carcinoma. Oncol. Lett. 2017, 14, 6735–6743. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.S.; Yang, M.H.; Liu, C.Y.; Liu, K.W.; Yang, T.T.; Chou, T.Y.; Hwang, T.Z.; Hsu, C.T. Decreasing Cytokeratin 17 Expression in Head and Neck Cancer Predicts Nodal Metastasis and Poor Prognosis: The First Evidence. Clin. Otolaryngol. 2018, 43, 1010–1018. [Google Scholar] [CrossRef]
- Tojyo, I.; Shintani, Y.; Nakanishi, T.; Okamoto, K.; Hiraishi, Y.; Fujita, S.; Enaka, M.; Sato, F.; Muragaki, Y. PD-L1 Expression Correlated with P53 Expression in Oral Squamous Cell Carcinoma. Maxillofac. Plast. Reconstr. Surg. 2019, 41, 56. [Google Scholar] [CrossRef]
- Coelho, B.A.; Peterle, G.T.; Santos, M.; Agostini, L.P.; Maia, L.L.; Stur, E.; Silva, C.V.; Mendes, S.O.; Almança, C.C.; Freitas, F.V.; et al. Keratins 17 and 19 Expression as Prognostic Markers in Oral Squamous Cell Carcinoma. Genet. Mol. Res. 2015, 14, 15123–15132. [Google Scholar] [CrossRef]
- Wang, L.; Song, H.; Yang, S. Overexpression of Keratin17 Is Associated with Prognosis of Oral Cancer in the Chinese Population. 2020. [CrossRef]
- Liu, Z.B.; Wu, J.; Ping, B.; Feng, L.Q.; Di, G.H.; Lu, J.S.; Shen, K.W.; Shen, Z.Z.; Shao, Z.M. Basal Cytokeratin Expression in Relation to Immunohistochemical and Clinical Characterization in Breast Cancer Patients with Triple Negative Phenotype. Tumori 2009, 95, 53–62. [Google Scholar] [CrossRef]
- da Silva, J.L.; Rodrigues, F.R.; de Mesquita, G.G.; Fernandes, P.V.; Thuler, L.C.S.; de Melo, A.C. Triple-Negative Breast Cancer: Assessing the Role of Immunohistochemical Biomarkers on Neoadjuvant Treatment. Breast Cancer 2021, 13, 31–44. [Google Scholar] [CrossRef]
- Cho, E.Y.; Chang, M.H.; Choi, Y.L.; Lee, J.E.; Nam, S.J.; Yang, J.H.; Park, Y.H.; Ahn, J.S.; Im, Y.H. Potential Candidate Biomarkers for Heterogeneity in Triple-Negative Breast Cancer (TNBC). Cancer Chemother. Pharmacol. 2011, 68, 753–761. [Google Scholar] [CrossRef]
- Dogu, G.G.; Ozkan, M.; Ozturk, F.; Dikilitas, M.; Er, O.; Ozturk, A. Triple-Negative Breast Cancer: Immunohistochemical Correlation with Basaloid Markers and Prognostic Value of Survivin. Med. Oncol. 2010, 27, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Kraus, J.A.; Beriwal, S.; Dabbs, D.J.; Ahrendt, G.M.; McGuire, K.P.; Johnson, R.R.; Badve, P.; Puhalla, S.L.; Bhargava, R. Predictors of Pathologic Complete Response After Standard Neoadjuvant Chemotherapy in Triple-Negative Breast Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2012, 20, 334–339. [Google Scholar] [CrossRef]
- Kawalerski, R.R.; Goncalves, M.T.; Pan, C.H.; Tseng, R.; Roa-Pena, L.; Leiton, C.V.; Torre-Healy, L.A.; Boyle, T.; Chowdhury, S.; Snider, N.T.; et al. Disassembly of Embryonic Keratin Filaments Promotes Pancreatic Cancer. bioRxiv 2022, 2022–2208. [Google Scholar] [CrossRef]
- Li, W.; Li, T.; Sun, C.; Du, Y.; Chen, L.; Du, C.; Shi, J.; Wang, W. Identification and Prognostic Analysis of Biomarkers to Predict the Progression of Pancreatic Cancer Patients. Mol. Med. 2022, 28, 43. [Google Scholar] [CrossRef]
- Lu, Y.; Li, D.; Liu, G.; Xiao, E.; Mu, S.; Pan, Y.; Qin, F.; Zhai, Y.; Duan, S.; Li, D.; et al. Identification of Critical Pathways and Potential Key Genes in Poorly Differentiated Pancreatic Adenocarcinoma. Onco Targets Ther. 2021, 14, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.; Thibodeau, B.J.; Baschnagel, A.M.; Fortier, L.E.; Kelley, Z.; Almradi, A.; Wilson, G.D.; Jury, R.P. Can Gene Expression Profiling Identify Pancreatic Ductal Adenocarcinoma Patients with Short or Long-Term Prognosis? J. Pancreas 2018, 19, 118–125. [Google Scholar]
- Wang, J.; Lan, L.; Ma, B.; Ren, G.; Yin, C. KRT17 Accelerates Cell Proliferative and Invasive Potential of Laryngeal Squamous Cell Carcinoma (LSCC) through Regulating AKT/mTOR and Wnt/β-Catenin Pathways. Evid. Based Complement. Altern. Med. 2022, 2022, 6176043. [Google Scholar] [CrossRef]
- Hashiguchi, M.; Masuda, M.; Kai, K.; Nakao, Y.; Kawaguchi, A.; Yokoyama, M.; Aishima, S. Decreased Cytokeratin 7 Expression Correlates with the Progression of Cervical Squamous Cell Carcinoma and Poor Patient Outcomes. J. Obs. Gynaecol. Res. 2019, 45, 2228–2236. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Ji, Y.; Ma, L.; Ge, S.; Chen, J.; Wu, S.; Huang, T.; Sheng, Y.; Wang, L.; Yi, N. Keratin 17 Upregulation Promotes Cell Metastasis and Angiogenesis in Colon Adenocarcinoma. Bioengineered 2021, 12, 12598–12611. [Google Scholar] [CrossRef]
- Ujiie, D.; Okayama, H.; Saito, K.; Ashizawa, M.; Min, A.K.T.; Endo, E.; Kase, K.; Yamada, L.; Kikuchi, T.; Hanayama, H.; et al. KRT17 as a Prognostic Biomarker for Stage II Colorectal Cancer. Carcinogenesis 2020, 41, 591–599. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, S.; Ye, S.; Shen, Z.; Gao, L.; Han, Z.; Zhang, P.; Luo, F.; Chen, S.; Kang, M. Keratin 17 Activates AKT Signalling and Induces Epithelial-Mesenchymal Transition in Oesophageal Squamous Cell Carcinoma. J. Proteom. 2020, 211, 103557. [Google Scholar] [CrossRef]
- Haye, K.; Babu, S.; Oblein, L.; Gupta, R.; Akalin, A.; Escobar-Hoyos, L.F.; Shroyer, K.R. Keratin 17 Expression Predicts Poor Clinical Outcome in Patients with Advanced Esophageal Squamous Cell Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.; Tittarelli, A.; Paillaleve, N.; Del Pozo, M.; Rojas-Sepulveda, D.; Barria, O.; Fluxa, P.; Hott, M.; Martin, C.; Quezada, C.; et al. The Evaluation of 17 Gastrointestinal Tumor Markers Reveals Prognosis Value for MUC6, CK17, and CD10 in Gallbladder-Cancer Patients. Diagnostics 2021, 11, 153. [Google Scholar] [CrossRef]
- Kim, K.; Lee, H.W.; Chae, S.W.; Kim, D.-H.; Do, I.G.; Lee, H.J.; Do, S.-I.; Min, K.-W.; Pyo, J.-S.; Shin, J.-H. Cytokeratin 17 Expression Is Associated with Poor Prognosis in Gallbladder Adenocarcinoma. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xu, D.H.; Huang, X.X.; Zhu, C.C.; Xu, J.; Zhang, Z.Z.; Zhao, G. Keratin17 Promotes Tumor Growth and Is Associated with Poor Prognosis in Gastric Cancer. J. Cancer 2018, 9, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Alkhasawneh, A.; Duckworth, L.V.; George, T.J.; Desai, N.V.; Sommerfeld, A.J.; Lu, X.; Toro, T.Z. Clinical, Morphologic, and Immunophenotypic Characteristics of Ampullary Carcinomas with an Emphasis on SMAD4 Expression. J. Gastrointest. Oncol. 2016, 7, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Sarlos, D.P.; Yusenko, M.V.; Peterfi, L.; Szanto, A.; Kovacs, G. Dual Role of KRT17: Development of Papillary Renal Cell Tumor and Progression of Conventional Renal Cell Carcinoma. J. Cancer 2019, 10, 5124–5129. [Google Scholar] [CrossRef] [PubMed]
- Thike, A.A.; Iqbal, J.; Cheok, P.Y.; Chong, A.P.; Tse, G.M.; Tan, B.; Tan, P.; Wong, N.S.; Tan, P.H. Triple Negative Breast Cancer: Outcome Correlation with Immunohistochemical Detection of Basal Markers. Am. J. Surg. Pathol. 2010, 34, 956–964. [Google Scholar] [CrossRef]
- Wu, J.; Xu, H.; Ji, H.; Zhai, B.; Zhu, J.; Gao, M.; Zhu, H.; Wang, X. Low Expression of Keratin17 Is Related to Poor Prognosis in Bladder Cancer. Onco Targets Ther. 2021, 14, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Liu, W.; Yong, L.; Liu, D.; Lin, X.; Huang, Y.; Wang, H.; Cai, F. Reduced Expression of KRT17 Predicts Poor Prognosis in HER2(High) Breast Cancer. Biomolecules 2022, 12, 1183. [Google Scholar] [CrossRef]
- Liang, W.; Liu, H.; Zeng, Z.; Liang, Z.; Xie, H.; Li, W.; Xiong, L.; Liu, Z.; Chen, M.; Jie, H.; et al. KRT17 Promotes T-Lymphocyte Infiltration Through the YTHDF2-CXCL10 Axis in Colorectal Cancer. Cancer Immunol. Res. 2023, 11, 875–894. [Google Scholar] [CrossRef]
- Ingenwerth, M.; Nyirády, P.; Hadaschik, B.; Szarvas, T.; Reis, H. The Prognostic Value of Cytokeratin and Extracellular Collagen Expression in Urinary Bladder Cancer. Curr. Mol. Med. 2022, 22, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Dundr, P.; Bazalová, B.; Bártů, M.; Bosse, T.; Drozenová, J.; Fabian, P.; Fadare, O.; Hausnerová, J.; Jakša, R.; Laco, J. The Cytokeratin 17 Expression in Primary Ovarian Tumors Has Diagnostic but Not Prognostic Significance. Virchows Arch. 2022, 481, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Diallo, R.; Ting, E.; Gluz, O.; Herr, A.; Schütt, G.; Geddert, H.; Mohrmann, S.; Gabbert, H.E.; Nitz, U.; Poremba, C. C-Kit Expression in High-Risk Breast Cancer Subgroup Treated with High-Dose or Conventional Dose-Dense Chemotherapy. Verh. Dtsch. Ges. Pathol. 2006, 90, 177–185. [Google Scholar]
- Da Silva, M.A.C.N.; Manhães, V.P.R.; Gasparotto Júnior, L.; Tsukumo, D.M.L.; Lalli, C.A. Pancytopenia as an Initial Manifestation of Prostate Cancer: A Case Report. J. Med. Case Rep. 2021, 15, 247. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, J.; Li, H.; Wang, X.; Yang, Q.; Shao, Y. Keratin 17 Is a Prognostic Biomarker in Endometrial Carcinoma and Correlates with Immune Invasion. 2022. [Google Scholar] [CrossRef]
- Zhou, L.; Lu, H.; Zeng, F.; Zhou, Q.; Li, S.; Wu, Y.; Yuan, Y.; Xin, L. Constructing a New Prognostic Signature of Gastric Cancer Based on Multiple Data Sets. Bioengineered 2021, 12, 2820–2835. [Google Scholar] [CrossRef]
- Luo, J.; Du, X. A Promising Prognostic Signature for Lung Adenocarcinoma (LUAD) Patients Basing on 6 Hypoxia-Related Genes. Medicine 2021, 100, E28237. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, M.-Q.; Lei, L.; Fei, L.-R.; Zheng, Y.-W.; Huang, W.-J.; Li, Z.-H.; Liu, C.-C.; Xu, H.-T. Overexpression of KRT17 Promotes Proliferation and Invasion of Non-Small Cell Lung Cancer and Indicates Poor Prognosis. Cancer Manag. Res. 2019, 11, 7485–7497. [Google Scholar] [CrossRef]
- Han, W.; Hu, C.; Fan, Z.J.; Shen, G.L. Transcript Levels of Keratin 1/5/6/14/15/16/17 as Potential Prognostic Indicators in Melanoma Patients. Sci. Rep. 2021, 11, 1023. [Google Scholar] [CrossRef]
- Miñoza, J.M.A.; Rico, J.A.; Zamora, P.R.F.; Bacolod, M.; Laubenbacher, R.; Dumancas, G.G.; de Castro, R. Biomarker Discovery for Meta-Classification of Melanoma Metastatic Progression Using Transfer Learning. Genes 2022, 13, 2303. [Google Scholar] [CrossRef] [PubMed]
- Wach, S.; Taubert, H.; Weigelt, K.; Hase, N.; Köhn, M.; Misiak, D.; Hüttelmaier, S.; Stöhr, C.G.; Kahlmeyer, A.; Haller, F.; et al. RNA Sequencing of Collecting Duct Renal Cell Carcinoma Suggests an Interaction between miRNA and Target Genes and a Predominance of Deregulated Solute Carrier Genes. Cancers 2019, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- Modi, A.; Purohit, P.; Gadwal, A.; Ukey, S.; Roy, D.; Fernandes, S.; Banerjee, M. In-Silico Analysis of Differentially Expressed Genes and Their Regulating microRNA Involved in Lymph Node Metastasis in Invasive Breast Carcinoma. Cancer Investig. 2022, 40, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Kawaguchi, A.; Fukai, J.; Iwadate, Y.; Kajiwara, K.; Hondoh, H.; Yamanaka, R. Survival Prediction Based on the Gene Expression Associated with Cancer Morphology and Microenvironment in Primary Central Nervous System Lymphoma. PLoS ONE 2021, 16, e0251272. [Google Scholar] [CrossRef]
- Li, C.; Teng, Y.; Wu, J.; Yan, F.; Deng, R.; Zhu, Y.; Li, X. A Pan-Cancer Analysis of the Oncogenic Role of Keratin 17 (KRT17) in Human Tumors. Transl. Cancer Res. 2021, 10, 4489–4501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Feng, Z.; Lu, L.; Li, Y.; Liu, Y.; Chen, Y. Analysis of the Expression and Role of Keratin 17 in Human Tumors. Front. Genet. 2022, 13, 801698. [Google Scholar] [CrossRef]
- Li, M.; Rao, X.; Cui, Y.; Zhang, L.; Li, X.; Wang, B.; Zheng, Y.; Teng, L.; Zhou, T.; Zhuo, W. The Keratin 17/YAP/IL6 Axis Contributes to E-Cadherin Loss and Aggressiveness of Diffuse Gastric Cancer. Oncogene 2022, 41, 770–781. [Google Scholar] [CrossRef]
- Pan, C.H.; Otsuka, Y.; Sridharan, B.; Woo, M.; Leiton, C.V.; Babu, S.; Goncalves, M.T.; Kawalerski, R.R.; Bai, J.D.K.; Chang, D.V.K.; et al. An Unbiased High-Throughput Drug Screen Reveals a Potential Therapeutic Vulnerability in the Most Lethal Molecular Subtype of Pancreatic Cancer. Mol. Oncol. 2020, 14, 1800–1816. [Google Scholar] [CrossRef]
- Anderson, L.; Seilhamer, J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 1997, 18, 533–537. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Brockmann, R.; Beyer, A.; Heinisch, J.J.; Wilhelm, T. Posttranscriptional Expression Regulation: What Determines Translation Rates? PLoS Comput. Biol. 2007, 3, e57. [Google Scholar] [CrossRef]
- Marguerat, S.; Schmidt, A.; Codlin, S.; Chen, W.; Aebersold, R.; Bähler, J. Quantitative Analysis of Fission Yeast Transcriptomes and Proteomes in Proliferating and Quiescent Cells. Cell 2012, 151, 671–683. [Google Scholar] [CrossRef]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global Quantification of Mammalian Gene Expression Control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Claydon, A.J.; Beynon, R. Proteome Dynamics: Revisiting Turnover with a Global Perspective. Mol. Cell. Proteom. MCP 2012, 11, 1551–1565. [Google Scholar] [CrossRef] [PubMed]
- Kerr, D.J.; Yang, L. Personalising Cancer Medicine with Prognostic Markers. eBioMedicine 2021, 72. [Google Scholar] [CrossRef] [PubMed]
- Lozar, T.; Laklouk, I.; Golfinos, A.E.; Gavrielatou, N.; Xu, J.; Flynn, C.; Keske, A.; Yu, M.; Bruce, J.Y.; Wang, W.; et al. Stress Keratin 17 Is a Predictive Biomarker Inversely Associated with Response to Immune Check-Point Blockade in Head and Neck Squamous Cell Carcinomas and Beyond. Cancers 2023, 15, 4905. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The Immune Contexture and Immunoscore in Cancer Prognosis and Therapeutic Efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, R.P.; DePianto, D.J.; Jacob, J.T.; Han, M.C.; Chung, B.M.; Batazzi, A.S.; Poll, B.G.; Guo, Y.; Han, J.; Ong, S.; et al. Keratin-Dependent Regulation of Aire and Gene Expression in Skin Tumor Keratinocytes. Nat. Genet. 2015, 47, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Karin, N. CXCR3 Ligands in Cancer and Autoimmunity, Chemoattraction of Effector T Cells, and Beyond. Front. Immunol. 2020, 11, 541785. [Google Scholar] [CrossRef] [PubMed]
- House, I.G.; Savas, P.; Lai, J.; Chen, A.X.; Oliver, A.J.; Teo, Z.L.; Todd, K.L.; Henderson, M.A.; Giuffrida, L.; Petley, E.V.; et al. Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Oldenhuis, C.N.A.M.; Oosting, S.F.; Gietema, J.A.; De Vries, E.G.E. Prognostic versus Predictive Value of Biomarkers in Oncology. Eur. J. Cancer 2008, 44, 946–953. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Scheel, A.H.; Dietel, M.; Heukamp, L.C.; Jöhrens, K.; Kirchner, T.; Reu, S.; Rüschoff, J.; Schildhaus, H.-U.; Schirmacher, P.; Tiemann, M.; et al. Harmonized PD-L1 Immunohistochemistry for Pulmonary Squamous-Cell and Adenocarcinomas. Mod. Pathol. 2016, 29, 1165–1172. [Google Scholar] [CrossRef]
- Munn, Z.; Peters, M.D.J.; Stern, C.; Tufanaru, C.; McArthur, A.; Aromataris, E. Systematic Review or Scoping Review? Guidance for Authors When Choosing between a Systematic or Scoping Review Approach. BMC Med. Res. Methodol. 2018, 18, 143. [Google Scholar] [CrossRef] [PubMed]
| N (%) | |
|---|---|
| K17 evaluation method | |
| Protein | 33 (56.9%) |
| RNA | 20 (34.5%) |
| RNA and Protein | 5 (8.6%) |
| Cancer type | |
| Breast | 11 (19.0%) |
| Gastrointestinal | 19 (32.8%) |
| Genitourinary | 5 (8.6%) |
| Gynecologic | 8 (13.8%) |
| Head and Neck | 8 (13.8%) |
| Lung | 2 (3.4%) |
| Lymphoma | 1 (1.7%) |
| Skin | 2 (3.4%) |
| Pan-cancer | 2 (3.4%) |
| HPV-related cancers | |
| Oropharyngeal HNSCC | 2 (3.4%) |
| Non-oropharyngeal/any HNSCC | 6 (10.3%) |
| Cervical SCC | 2 (3.4%) |
| Cervical, any | 1 (1.7%) |
| Study design | |
| Retrospective | 44 (75.9%) |
| Prospective | 8 (13.8%) |
| Online database/computational | 6 (10.3%) |
| Study objective | |
| K17 only | 48 (82.8%) |
| K17 in a gene signature | 4 (6.8%) |
| K17 in a protein panel | 6 (10.3%) |
| Clinical significance | |
| Prognostic | 54 (93.1%) |
| Predictive | 4 (6.9%) |
| immunotherapy | 2 (3.4%) |
| chemotherapy | 2 (3.4%) |
| Cancer Type | Evaluation Method | N (All Studies) | N (Negative Correlation) | N (Positive Correlation) | N (No Correlation) |
|---|---|---|---|---|---|
| Bladder | Protein | 2 | - | 1 | 1 |
| Breast | Protein | 3 | 1 | 2 | 1 * |
| RNA | 2 | - | 2 | - | |
| Cervical | Protein | 4 | 3 | - | 1 |
| Colorectal | Protein | 3 | 2 | 1 | - |
| Endometrial | Protein | 1 | 1 | - | - |
| RNA | 2 | 2 | - | - | |
| Esophageal | Protein | 2 | 2 | - | - |
| RNA | 1 | 1 | - | - | |
| Gallbladder | Protein | 2 | 2 | - | - |
| Gastric | Protein | 3 | 3 | - | - |
| RNA | 2 | 1 | 1 | - | |
| HNSCC | Protein | 5 | 2 | 2 | 1 |
| RNA | 3 | 3 | - | - | |
| Lung | RNA | 2 | 2 | - | - |
| Lymphoma | RNA | 1 | - | - | 1 |
| Ovarian | Protein | 2 | 1 | - | 1 |
| Melanoma | RNA | 2 | 2 | - | - |
| Pancreatic | Protein | 3 | 3 | - | - |
| RNA | 4 | 3 | - | 1 | |
| Renal | Protein | 1 | 1 | - | - |
| RNA | 1 | 1 | - | - | |
| TNBC | Protein | 7 | 2 | 5 | - |
| RNA | 1 | 1 | - | - | |
| Urothelial ** | Protein | 1 | - | 1 | - |
| Author, Year (Ref) | Cancer Type | Evaluation Method | N | %K17 High Expressors | Endpoint | Correlation between K17 and Outcome | Effect Size |
|---|---|---|---|---|---|---|---|
| Wu (2021) [64] | Bladder | IHC; TMA | 101 | 55% | OS | positive | Univariate HR (high): 0.11, p < 0.01 Multivariate HR (low): 4.263; p = 0.019 |
| Ingenwerth (2022) [67] | Bladder | IHC | 190 | 54% | DSS | no correlation | NR |
| Rodriguez-Pinilla (2007) [37] | Breast | IHC; TMA | 245 | 12% | MFS, OS | negative | Univariate RR (high): 1.22, p = NS for MFS and HR = 1.8, p = NS for OS |
| Tang (2022) [65] | Breast | IHC; Transcriptome | 150 | NR | OS | positive in HER2high; inverse in HER2low | Univariate HR (low): 0.6, p = 0.002 |
| He (2021) [34] * | Cervical | ELISA | 134 | NR | OS | negative | NR |
| Mockler (2017) [23] | Cervical adenocarcinoma | IHC | 90 | <40%: 64% >90%: 13% | OS | negative | Univariate HR (high): 3.47, p = 0.013 Multivariate HR (high): 2.76, p = 0.048 |
| Escobar-Hoyos (2014) [22] | Cervical SCC | IHC; TMA | 65 | 35% | OS | negative | NR |
| Hashiguchi (2019) [53] | Cervical SCC | IHC | 129 | 60% | OS | no correlation | Univariate HR (high): 0.56, p = NS Multivariate HR: 0.65, p = 0.3 |
| Ji (2021) [54] | Colon adenocarcinoma | IHC | 78 | 50% | Survival state | negative | NR |
| Ujiie (2020) [55] | Colorectal | IHC; Transcriptome; TMA | 154 (Cohort 1 = 110, Cohort 2 = 44) | 50% | RFS | negative | Univariate HR (high): 5.30, p < 0.001 Multivariate HR (high): 7.81, p < 0.001 |
| Liang (2023) [66] | Colorectal | IHC | 446 | NR | OS, recurrence rate | positive | Univariate HR (high): 0.33; p = 0.0004 for OS and HR = 0.33, p < 0.01 for DFS |
| Bai (2019) [30] | Endometrial | IHC; Transcriptome | 119 | 39% | OS | negative | Univariate HR (high): 1.75, p = 0.049 Multivariate HR (high): 2.0, p = 0.019 |
| Wang (2013) [27] | Epithelial ovarian cancer | IHC | 104 | 54% | OS | negative | Univariate RR (high): 1.44, p < 0.01 Multivariate RR (high): 1.531, p < 0.01 |
| Liu (2020) [56] | Esophageal SCC | IHC; TMA | 64 | 66% | OS | negative | Univariate HR (high): 2.51, p = 0.045 Multivariate HR (high): 5.383, p = 0.012 |
| Haye (2021) [57] | Esophageal SCC | IHC; Transcriptome | 68 | 58% | EFS | negative | Univariate HR (high, advanced stages subgroup): 2.08, p = 0.0384. |
| Carrasco (2021) [58] | Gallbladder | IHC; TMA | 162 | 73% | OS | negative | Univariate HR (high, poorly differentiated): 2.0, p < 0.01, Multivariate HR (high, poorly differentiated): 2.46, p = 0.037 |
| Kim (2017) [59] | Gallbladder adenocarcinoma | IHC; TMA | 77 | 53% | DSS | negative | Univariate HR (high): 4.76, p = 0.001 Multivariate HR (high): 3.62, p = 0.01 |
| Alkhasawneh (2016) [61] | Gastric | IHC | 63 | 51% | OS | negative | NR |
| Hu (2018) [60] | Gastric | IHC; TMA | 569 | 56% | OS | negative | Univariate HR (high): 1.454, p < 0.01 Multivariate HR (high): 1.336, p < 0.01 |
| Ide (2012) [26] | Gastric adenocarcinoma | IHC; TMA | 192 | 50% | 5-year DSS | negative | Univariate HR (low): 0.51, p = 0.004 Multivariate HR (low): 0.786, p = 0.049 |
| Xu (2018) [39] | HNSCC | IHC; TMA | 106 | 54% | DSS | positive | Univariate HR (low): 2.04, p = 0.022 Multivariate HR (low): 1.854, p = NS |
| Wang (2022) [15] | HNSCC | IF | 107 | 70% | OS, DCR | negative | NR |
| Coelho (2015) [41] | HNSCC, oral | IHC; TMA | 67 | 79% | DFS, DSS | positive | Multivariate HR (low) in subgroup treated with surgery and RT: 4.11; p = 0.027 for DFS and 4.75; p = 0.016 for DSS |
| Tojyo (2019) [40] | HNSCC, oral | IHC | 49 | 20% | 5-year DFS | no correlation | NR |
| Regenbogen (2018) [24] | HNSCC | IHC | 78 | 96% | survival state, OS | negative | Multivariate HR (high): 2.30, p = 0.0152 |
| Dundr (2022) [68] | Ovarian | IHC; TMA | 125 | 14% | OS, DFS, LFS, MFS | no correlation | NR |
| Roa-Peña (2019) [31] | Pancreatic | IHC; Transcriptome | 74 | 24% | OS | negative | Univariate HR (high): 2.96, p = 0.008 Multivariate HR (high): 3.09, p = 0.015 |
| Roa-Peña (2021) [35] ** | Pancreatic | IHC | 211 *** | 35% | OS | negative | Univariate HR (high, combined cohorts): 1.7, p = 0.0017, Multivariate HR (high, discovery set): 1.9, p = 0.0235 |
| Kawalerski (2022) [48] | Pancreatic | IHC; Mass spec | 26 | 23% | OS | negative | Univariate HR (highly detergent-soluble): 2.854; P = 0.046 |
| Sarlos (2019) [62] | Renal cell | IHC; TMA | 692 | 14% | DSS, recurrence | negative | RR (high) for post-operative recurrence: 2.5, p < 0.01 |
| Liu (2009) [43] | TNBC | IHC | 112 | 34% | RFS, OS | negative | Univariate HR (high): RFS: HR = 2.121, p = NS for RFS and HR = 2.142, p= 0.037 for OS Multivariate: HR = 0.910, p = NS for RFS and HR= 0.933, p = NS for OS |
| Dogu (2010) [46] | TNBC | IHC | 33 | 74% | DFS | no correlation | NR |
| Thike (2010) [63] | TNBC | IHC; TMA | 653 | NR | DFS | negative | NR |
| Cho (2011) [45] | TNBC | IHC | 88 | 53% | RFS < 36 months | no correlation | NR |
| Kraus (2012) [47] | TNBC | IHC; TMA | 56 | 45% | PCR | no correlation | NR |
| Merkin (2017) [29] | TNBC | IHC; Transcriptome | 149 | >0%: 82% | EFS | no correlation | NR |
| daSilva (2021) [70] | TNBC | IHC; TMA | 168 | 91% | EFS, OS | no correlation | NR |
| Langner (2004) [36] | Urothelial | IHC; TMA | 53 | 40% | MFS | positive | NR |
| Study ID (Ref) | Cancer Type | Data Derived from | Sample Size | K17 Cut-Off | Endpoint | Correlation between K17 and Outcome | Effect Size |
|---|---|---|---|---|---|---|---|
| Tang (2022) [65] | Breast | KM Plotter | NR | Not specified | OS | Positive | HR (high): 0.60, p = 0.002 |
| Modi (2022) [78] | Breast | KM Plotter | 1070 | 50% | OS | Positive | Univariate HR (high): 0.59, p = 0.0017 |
| Bai (2019) [30] | Endometrial | TCGA | 271 | NR, determined experimentally, 39% K17 high | OS | negative | Univariate HR (high): 1.8, p = 0.01, Multivariate HR (high): 1.4, p = 0.086 |
| Zhang (2022) [71] | Endometrial | TCGA | 552 | Median | OS | negative | Univariate HR (high): 1.65, p = 0.018, Multivariate HR (high): 1.29, p = 0.409 |
| Haye (2021) [57] | Esophageal SCC | TCGA | 90 | NR, determined experimentally, 58% K17 high | EFS | negative | Univariate HR (high): 2.17, p = 0.04, Multivariate HR(high): 2.4, p = 0.04 |
| Zhou (2021) [72] | Gastric | KM Plotter | 635 | NR, determined experimentally | OS | negative | Univariate HR (high, validation cohort): 1.59, p < 0.01 |
| Li (2022) [82] | Gastric cancer | TCGA | diffuse: 135, intestinal: 146 | 50% | OS | Positive | NR |
| Wang (2020) [42] | HNSCC, oral | in study (qRT-PCR) | 135 | 50% | OS | negative | Multivariate HR (high): 2.49 p = 0.004 |
| Wang (2022) [52] | HNSCC, laryngeal | in study | 42 | Median | OS | negative | NR |
| Kitamura (2017) [38] | HNSCC, oral | in study (PBMC) | 19 | 32%, determined experimentally | DFS | negative | NR |
| Luo (2021) [73] | Lung adenocarcinoma | TCGA | 500 | NR, determined experimentally | OS | negative | Multivariate HR (high): 1.1, p = 0.028 |
| Wang (2019) [74] | Lung; NSCLC | UALCAN, KM Plotter | NR | Not specified | OS | negative (KM Plotter cohort only) | Univariate HR (high): 1.45, p = 1.1 × 10−8 |
| Han (2021) [75] | Melanoma | TCGA, GEO | 458 | 50% | OS | negative | Univariate HR (high): 1.5, p = 0.0018 |
| Miñoza (2022) [76] | Melanoma | TCGA | 468 | 50% | OS | negative | NR |
| Li (2021) [80] | Pan-cancer | TCGA, GEO | NR | 50% | OS, DFS | negative | For OS: SKCM: n = 515; HR (high, OS) 1.4, p = 0.038; PAAD: 364; HR 1.5; p = 0.017; mesothelioma 82; HR 3.1, p = 2.3 × 10−5; LUAD: 178, HR 1.5, p = 0.038; LIHC: 478, HR 1.6, p = 0.0033; RCC: 458, HR 1.5; p = 0.0018. For DFS: mesothelioma: 82, HR (high) 1.8; p = 0.048; PAAD: 178, HR 1.7, p = 0.017 |
| Zhang (2022) [71] | Pan-cancer | KM Plotter | NR | Median | OS, RFS | negative and positive | For OS: RCC: HR (high) 1.64, p = 0.003, LIHC: HR 1.63, p = 0.005, LUAD: HR 1.53, p = 0.007, PAAD: HR 1.99, p = 0.008, UCEC: HR 1.89, p = 0.004, BC: HR (high) 0.55, p < 0.001. For RFS: THCA: HR 0.41, p = 0.04, RCC: HR 0.15, p = 0.034, UCEC: HR 0.5, p = 0.008, BLCA: HR 2.19, p = 0.026, PAAD: HR 2.61, p = 0.018 |
| Roa-Peña (2019) [31] | Pancreatic | TCGA, prior literature | Cohort 1: 145, Cohort 2: 124 | 76%, determined experimentally | OS | negative | Univariate HR (high, combined): 1.69, p = 0.003 Multivariate HR (high): 1.79, p = 0.037 |
| Li (2022) [49] | Pancreatic | TCGA | 178 | Median | OS | No correlation | NR |
| Lu (2021) [50] | Pancreatic adenocarcinoma | TCGA | 170 | Median TPM | OS | negative | Univariate HR (high): 1.15, p = 0.018 |
| Stone (2018) [51] | Pancreatic adenocarcinoma | in study | 24 | Median | OS | negative | NR |
| Takashima (2021) [79] | Primary CNS lymphoma | in study | 31 | 50% | OS | No correlation | NR |
| Wach (2019) [77] | Renal | TCGA | ccRCC n = 522; pRCC: 284 | Median | OS | negative | NR |
| Merkin (2017) [29] | TNBC | TCGA | 149 (IDC only) | NR, determined experimentally | EFS | negative (subgroup only) | NR |
| Study ID | Cancer Type | Study Type | Study Design | K17 Evaluation Method | Treatment | Sample Size | % K17 High | Endpoint | Correlation between K17 and Outcome | Effect Size |
|---|---|---|---|---|---|---|---|---|---|---|
| Diallo (2006) [69] | Breast | Protein | P | IHC; TMA | High-dose (HDCT) vs. Dose dense chemotherapy (DDCT) | 224 | 6% | OS | negative | Univariate HR (high, DDCT arm): 5.1, p < 0.001 |
| Liang (2023) [66] | Colorectal | Protein | R | IHC | anti PD-1 in dMMR patients | 30 | NR | objective response to therapy (RECIST) | positive | NR |
| Wang (2022) [15] | HNSCC | Protein | R | IHC; TMA | Pembrolizumab | 26 | 70% | DCR, PFS, OS | negative | NR |
| Pan (2020) [83] | Pancreatic | RNA | R | Transcriptome (APGI) | Gemcitabine | 94 | 24% | OS | negative | Univariate HR (high,): 1.8, p = 0.335, Multivariate HR: 1.79, p = 0.046 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozar, T.; Wang, W.; Gavrielatou, N.; Christensen, L.; Lambert, P.F.; Harari, P.M.; Rimm, D.L.; Burtness, B.; Grasic Kuhar, C.; Carchman, E.H. Emerging Prognostic and Predictive Significance of Stress Keratin 17 in HPV-Associated and Non HPV-Associated Human Cancers: A Scoping Review. Viruses 2023, 15, 2320. https://doi.org/10.3390/v15122320
Lozar T, Wang W, Gavrielatou N, Christensen L, Lambert PF, Harari PM, Rimm DL, Burtness B, Grasic Kuhar C, Carchman EH. Emerging Prognostic and Predictive Significance of Stress Keratin 17 in HPV-Associated and Non HPV-Associated Human Cancers: A Scoping Review. Viruses. 2023; 15(12):2320. https://doi.org/10.3390/v15122320
Chicago/Turabian StyleLozar, Taja, Wei Wang, Niki Gavrielatou, Leslie Christensen, Paul F. Lambert, Paul M. Harari, David L. Rimm, Barbara Burtness, Cvetka Grasic Kuhar, and Evie H. Carchman. 2023. "Emerging Prognostic and Predictive Significance of Stress Keratin 17 in HPV-Associated and Non HPV-Associated Human Cancers: A Scoping Review" Viruses 15, no. 12: 2320. https://doi.org/10.3390/v15122320





