Congenital Zika Virus Infection Impairs Corpus Callosum Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Animals and ZIKV Infection
2.3. Virus Detection
2.4. Primary Neuronal Culture and Infection
2.5. Immunohistochemistry and Imaging
2.6. Immunocytochemistry and Quantitative Analyses
2.7. Histology
2.8. Dye Labelling
2.9. Isotropic Fractionator for Total and Neuronal Cell Counting
2.10. Statistical Analysis
3. Results
3.1. Congenital ZIKV Infection Leads to a Reduction in the Corpus Callosum in Mice
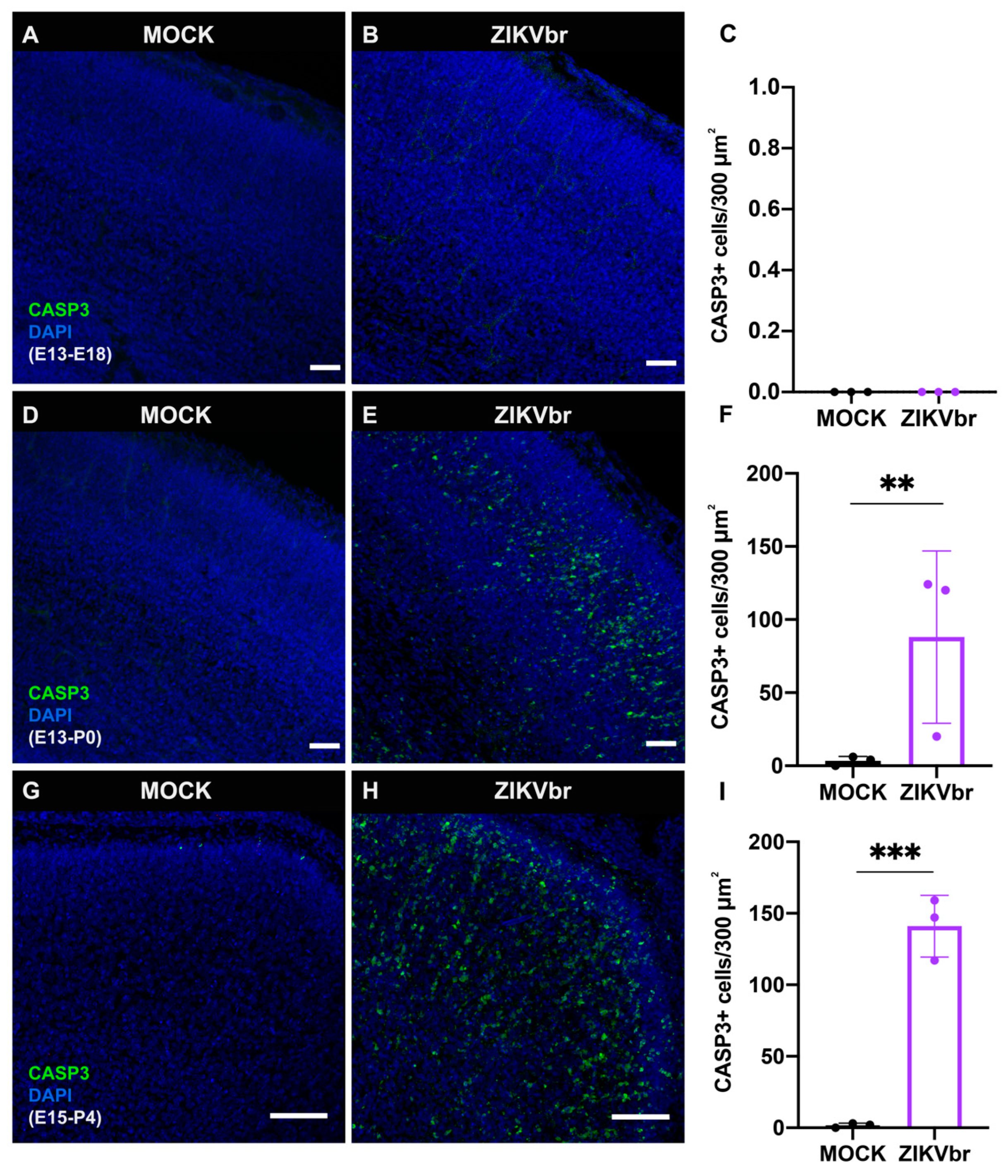
3.2. Congenital ZIKV Infection Causes Cell Death at the Cortical Plate
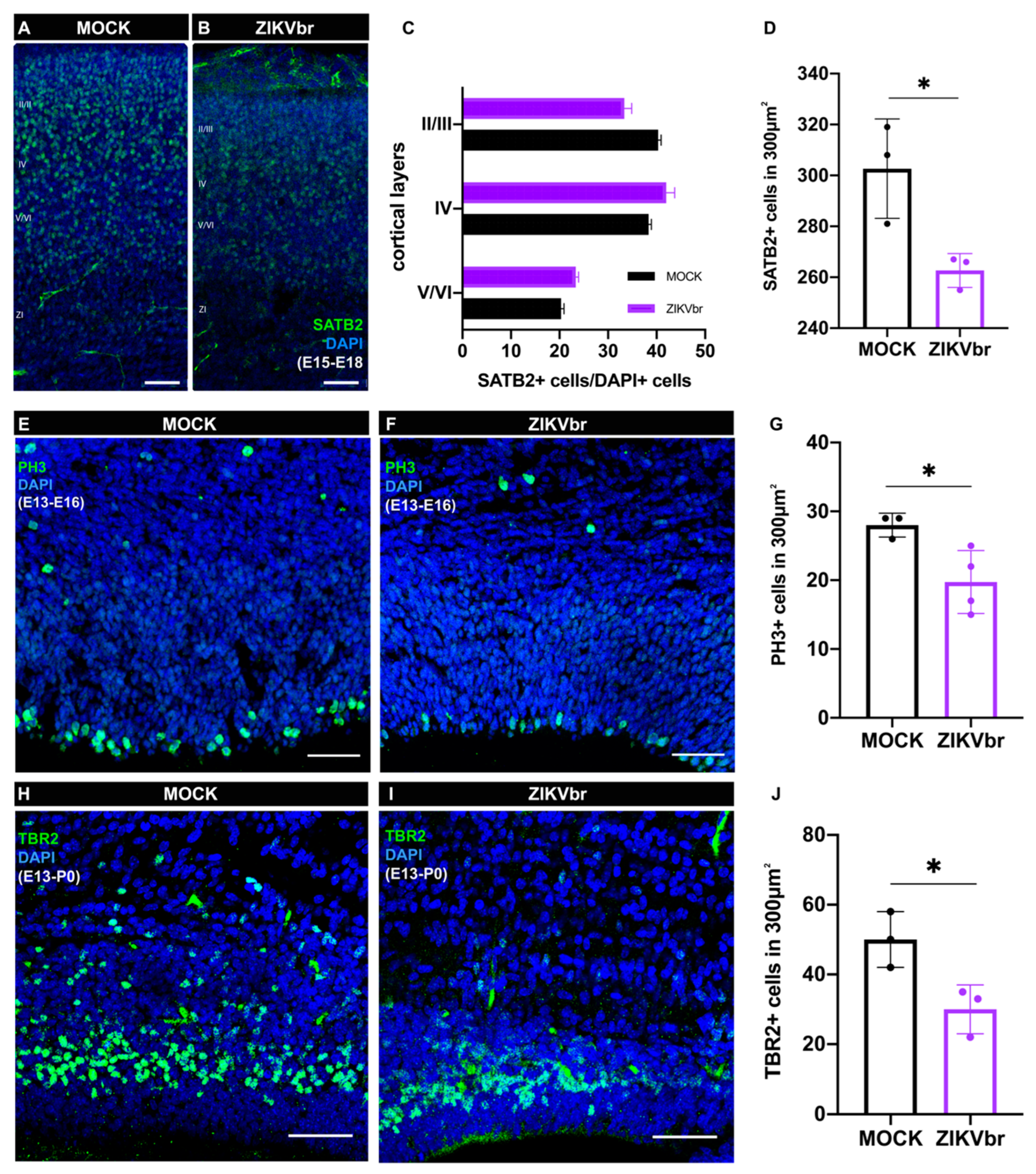
3.3. ZIKV Infection Leads to a Defect in Callosal Neuron Production
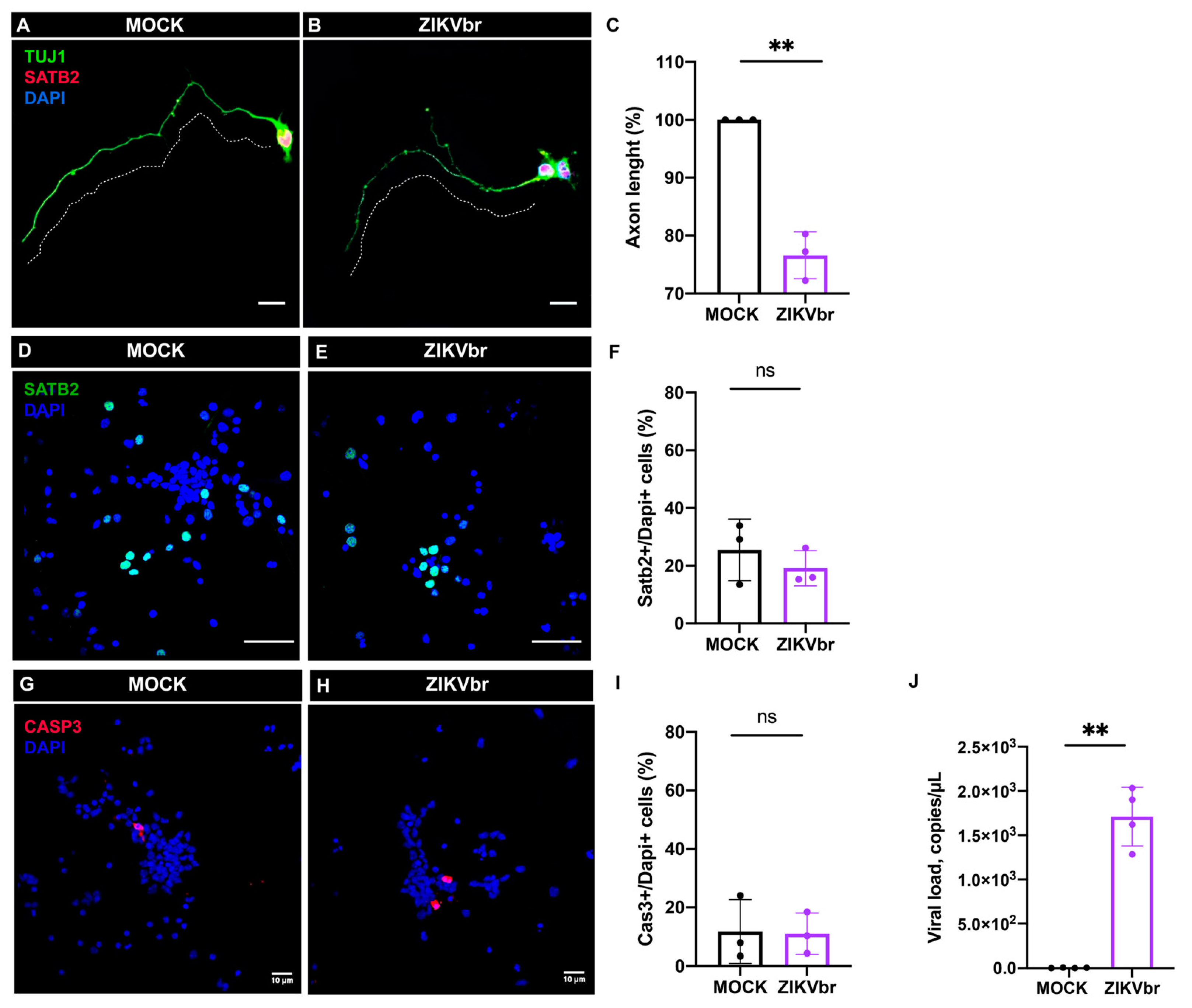
3.4. ZIKV Infection Impairs Axonal Growth of Callosal Neurons
3.5. ZIKV Infection Impairs Midline Glia Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.M.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Garcez, P.P.; Loiola, E.C.; Madeiro Da Costa, R.; Higa, L.M.; Trindade, P.; DelVecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Melo, A.S.; Aguiar, R.S.; Amorim, M.M.; Arruda, M.B.; Melo, F.O.; Ribeiro, S.T.; Batista, A.G.; Ferreira, T.; Dos Santos, M.P.; Sampaio, V.V.; et al. Congenital Zika virus infection: Beyond neonatal microcephaly. JAMA Neurol. 2016, 73, 1407–1416. [Google Scholar] [CrossRef]
- Chimelli, L.; Melo, A.S.O.; Avvad-Portari, E.; Wiley, C.A.; Camacho, A.H.S.; Lopes, V.S.; Machado, H.N.; Andrade, C.V.; Dock, D.C.A.; Moreira, M.E.; et al. The spectrum of neuropathological changes associated with congenital Zika virus infection. Acta Neuropathol. 2017, 133, 983–999. [Google Scholar] [CrossRef]
- De Fatima Vasco Aragao, M.; van der Linden, V.; Brainer-Lima, A.M.; Coeli, R.R.; Rocha, M.A.; Sobral da Silva, P.; Durce Costa Gomes de Carvalho, M.; van der Linden, A.; Cesario de Holanda, A.; Valenca, M.M. Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: Retrospective case series study. BMJ 2016, 353, i1901. [Google Scholar] [CrossRef]
- de Souza, A.S.; de Oliveira-Szjenfeld, P.S.; de Oliveira Melo, A.S.; de Souza, L.A.M.; Batista, A.G.M.; Tovar-Moll, F. Imaging findings in congenital Zika virus infection syndrome: An update. Child’s Nervous System. Child’s Nerv. Syst. 2018, 34, 85–93. [Google Scholar] [CrossRef]
- Tomasch, J. Size, distribution, and number of fibers in the human Corpus Callosum. Anat. Rec. 1954, 119, 119–135. [Google Scholar] [CrossRef]
- Tovar-Moll, F.; Moll, J.; De Oliveira-Souza, R.; Bramati, I.; Andreiuolo, P.A.; Lent, R. Neuroplasticity in human callosal dysgenesis: A diffusion tensor imaging study. Cereb Cortex 2007, 17, 531–541. [Google Scholar] [CrossRef]
- Tovar-Moll, F.; Monteiro, M.; Andrade, J.; Bramati, I.E.; Vianna-Barbosa, R.; Marins, T.; Rodrigues, E.; Dantas, N.; Behrens, T.E.; de Oliveira-Souza, R.; et al. Structural and functional brain rewiring clarifies preserved interhemispheric transfer in humans born without the corpus callosum. Proc. Natl. Acad. Sci. USA 2014, 111, 7843–7848. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.J.; Sherr, E.H.; Barkovich, A.J.; Richards, L.J. Clinical, genetic, and imaging findings identify new causes for corpus callosum development syndromes. Brain 2014, 137 Pt 6, 1579–1613. [Google Scholar] [CrossRef] [PubMed]
- Gobius, I.; Richards, L. Creating Connections in the Developing Brain: Mechanisms Regulating Corpus Callosum Development; Colloquium Series on The Developing Brain; Morgan & Claypool Publishers LLC: San Rafael, CA, USA, 2011; Volume 2, pp. 1–48. [Google Scholar]
- Shu, T.; Butz, K.G.; Plachez, C.; Gronostajski, R.M.; Richards, L.J. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J. Neurosci. 2003, 23, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Sundaresan, V.; McCarthy, M.M.; Richards, L.J. Slit2 guides both precrossing and postcrossing callosal axons at the midline in vivo. J. Neurosci. 2003, 23, 8176–8184. [Google Scholar] [CrossRef] [PubMed]
- Garcez, P.P.; Stolp, H.B.; Sravanam, S.; Christoff, R.R.; Ferreira, J.C.C.G.; Dias, A.A.; Pezzuto, P.; Higa, L.M.; Barbeito-Andrés, J.; Ferreira, R.O.; et al. Zika virus impairs the development of blood vessels in a mouse model of congenital infection. Sci. Rep. 2018, 8, 12774. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; An, S.; Chen, J.; Zhang, S.; Tan, C.; Yu, J.; Ye, H.; Wu, Y.; Yuan, J.; Wu, J.; et al. Neural progenitor cell pyroptosis contributes to zika virus-induced brain atrophy and represents a therapeutic target. Proc. Natl. Acad. Sci. USA 2020, 117, 23869–23878. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Q.; Jiang, Y.; Ye, Q.; Xu, D.; Gao, F.; Xu, J.W.; Wang, R.; Zhu, X.; Shi, L.; et al. Disruption of glial cell development by Zika virus contributes to severe microcephalic newborn mice. Cell Discov. 2018, 4, 43. [Google Scholar] [CrossRef]
- Lum, F.M.; Low, D.K.; Fan, Y.; Tan, J.J.; Lee, B.; Chan, J.K.; Rénia, L.; Ginhoux, F.; Ng, L.F. Zika virus infects human fetal brain microglia and induces inflammation. Clin. Infect. Dis. 2017, 64, 914–920. [Google Scholar] [CrossRef]
- van den Pol, A.N.; Mao, G.; Yang, Y.; Ornaghi, S.; Davis, J.N. Zika Virus Targeting in the Developing Brain. J. Neurosci. 2017, 37, 2161–2175. [Google Scholar] [CrossRef]
- Limonta, D.; Jovel, J.; Kumar, A.; Airo, A.M.; Hou, S.; Saito, L.; Branton, W.; Ka-Shu Wong, G.; Mason, A.; Power, C.; et al. Human fetal astrocytes infected with zika virus exhibit delayed apoptosis and resistance to interferon: Implications for persistence. Viruses 2018, 10, 646. [Google Scholar] [CrossRef]
- Ledur, P.F.; Karmirian, K.; Pedrosa, C.D.S.G.; Souza, L.R.Q.; Assis-de-Lemos, G.; Martins, T.M.; Ferreira, J.C.C.G.; de Azevedo Reis, G.F.; Silva, E.S.; Silva, D.; et al. Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci. Rep. 2020, 10, 1218. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Valério-Gomes, B.; Guimarães, D.M.; Szczupak, D.; Lent, R. The absolute number of oligodendrocytes in the adult mouse brain. Front. Neuroanat. 2018, 12, 90. [Google Scholar] [CrossRef] [PubMed]
- Barbeito-Andrés, J.; Pezzuto, P.; Higa, L.M.; Dias, A.A.; Vasconcelos, J.M.; Santos, T.M.P.; Ferreira, J.C.C.G.; Ferreira, R.O.; Dutra, F.F.; Rossi, A.D.; et al. Congenital Zika syndrome is associated with maternal protein malnutrition. Sci. Adv. 2020, 6, eaaw6284. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.F.; Xu, Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 2016, 19, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tan, Y.W.; Yam, W.K.; Tu, H.; Qiu, L.; Tan, E.K.; Chu, J.J.H.; Zeng, L. In utero infection of Zika virus leads to abnormal central nervous system development in mice. Sci. Rep. 2019, 9, 7298. [Google Scholar] [CrossRef]
- Shao, Q.; Herrlinger, S.; Yang, S.L.; Lai, F.; Moore, J.M.; Brindley, M.A.; Chen, J.F. Zika virus infection disrupts neurovascular development and results in postnatal microcephaly with brain damage. Development 2016, 143, 4127–4136. [Google Scholar] [CrossRef]
- Wu, K.Y.; Zuo, G.L.; Li, X.F.; Ye, Q.; Deng, Y.Q.; Huang, X.Y.; Cao, W.C.; Qin, C.F.; Luo, Z.G. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Research. Nat. Publ. Group 2016, 26, 645–654. [Google Scholar]
- Shi, Y.; Li, S.; Wu, Q.; Sun, L.; Zhang, J.; Pan, N.; Wang, Q.; Bi, Y.; An, J.; Lu, X.; et al. Vertical Transmission of the Zika Virus Causes Neurological Disorders in Mouse Offspring. Sci. Rep. 2018, 8, 3541. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, Y.; Dimos, J.T.; Fasano, C.A.; Phoenix, T.N.; Lemischka, I.R.; Ivanova, N.B.; Stifani, S.; Morrisey, E.E.; Temple, S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 2006, 9, 743–751. [Google Scholar] [CrossRef]
- Hevner, R.F. Intermediate progenitors and Tbr2 in cortical development. J. Anat. 2019, 235, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Doobin, D.J.; Dantas, T.J.; Vallee, R.B. Microcephaly as a cell cycle disease. Cell Cycle 2017, 16, 247–248. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, F.T.; Willard, K.A.; Wu, X.; Scoville, S.; Stice, S.L.; Brindley, M.A. Strain-dependent consequences of zika virus infection and differential impact on neural development. Viruses 2018, 10, 550. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.M.; Cowan, M.; Moonah, S.N.; Petri, W.A., Jr. The Impact of Systemic Inflammation on Neurodevelopment. Trends Mol. Med. 2018, 24, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Shan, C.; Dunn, T.J.; Xie, X.; Xia, H.; Gao, J.; Allende Labastida, J.; Zou, J.; Villarreal, P.P.; Schlagal, C.R.; et al. Role of Microglia in the Dissemination of Zika Virus from Mother to Fetal Brain. PloS Neglected Trop. Dis. 2020, 14, e0008413. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Jiang, Y.; Li, C.; Wang, Q.; Zhang, F.; Qin, C.F.; Wu, Q.F.; Li, J.; Xu, Z. Different Gene Networks Are Disturbed by Zika Virus Infection in A Mouse Microcephaly Model. Genom. Proteom. Bioinform. 2021, 18, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yue, X.; Shi, G.; Yue, Y.; Crockett, D.P.; Blair-Flynn, J.; Reuhl, K.; Tessarollo, L.; Zhou, R. Corpus Callosum Deficiency in Transgenic Mice Expressing Truncated Ephrin-A Receptor. J. Neurosci. 2003, 23, 10963–10970. [Google Scholar] [CrossRef]
- Mire, E.; Hocine, M.; Bazellières, E.; Jungas, T.; Davy, A.; Chauvet, S.; Mann, F. Developmental Upregulation of Ephrin-B1 Silences Sema3C/Neuropilin-1 Signaling during Post-crossing Navigation of Corpus Callosum Axons. Curr. Biol. 2018, 28, 1768–1782.e4. [Google Scholar] [CrossRef]
- Niquille, M.; Garel, S.; Mann, F.; Hornung, J.P.; Otsmane, B.; Chevalley, S.; Parras, C.; Guillemot, F.; Gaspar, P.; Yanagawa, Y.; et al. Transient neuronal populations are required to guide callosal axons: A role for semaphorin 3C. PLoS Biol. 2009, 7, e1000230. [Google Scholar] [CrossRef]
- Nielsen-Saines, K.; Brasil, P.; Kerin, T.; Vasconcelos, Z.; Gabaglia, C.R.; Damasceno, L.; Pone, M.; Abreu de Carvalho, L.M.; Pone, S.M.; Zin, A.A.; et al. Delayed childhood neurodevelopment and neurosensory alterations in the second year of life in a prospective cohort of ZIKV-exposed children. Nat. Med. 2019, 25, 1213–1217. [Google Scholar] [CrossRef]
- Moldrich, R.X.; Gobius, I.; Pollak, T.; Zhang, J.; Ren, T.; Brown, L.; Mori, S.; De Juan Romero, C.; Britanova, O.; Tarabykin, V.; et al. Molecular regulation of the developing commissural plate. J. Comp. Neurol. 2010, 518, 3645–3661. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christoff, R.R.; Quintanilha, J.H.; Ferreira, R.O.; Ferreira, J.C.C.G.; Guimarães, D.M.; Valério-Gomes, B.; Higa, L.M.; Rossi, Á.D.; Bellio, M.; Tanuri, A.; et al. Congenital Zika Virus Infection Impairs Corpus Callosum Development. Viruses 2023, 15, 2336. https://doi.org/10.3390/v15122336
Christoff RR, Quintanilha JH, Ferreira RO, Ferreira JCCG, Guimarães DM, Valério-Gomes B, Higa LM, Rossi ÁD, Bellio M, Tanuri A, et al. Congenital Zika Virus Infection Impairs Corpus Callosum Development. Viruses. 2023; 15(12):2336. https://doi.org/10.3390/v15122336
Chicago/Turabian StyleChristoff, Raissa Rilo, Jefferson H. Quintanilha, Raiane Oliveira Ferreira, Jessica C. C. G. Ferreira, Daniel Menezes Guimarães, Bruna Valério-Gomes, Luiza M. Higa, Átila D. Rossi, Maria Bellio, Amilcar Tanuri, and et al. 2023. "Congenital Zika Virus Infection Impairs Corpus Callosum Development" Viruses 15, no. 12: 2336. https://doi.org/10.3390/v15122336




