PDIA4 Is a Host Factor Important for Lymphocytic Choriomeningitis Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Viruses
2.3. Plasmids
2.4. Antibodies
2.5. Quantitative Proteomics Analysis
2.6. Western Blot
2.7. RNAi Experiments
2.8. Construction of Scramble-/PDIA4-/PDIA5-Knockout (KO) A549 Cells
2.9. Mouse Experiment
2.10. Statistical Analysis
3. Results
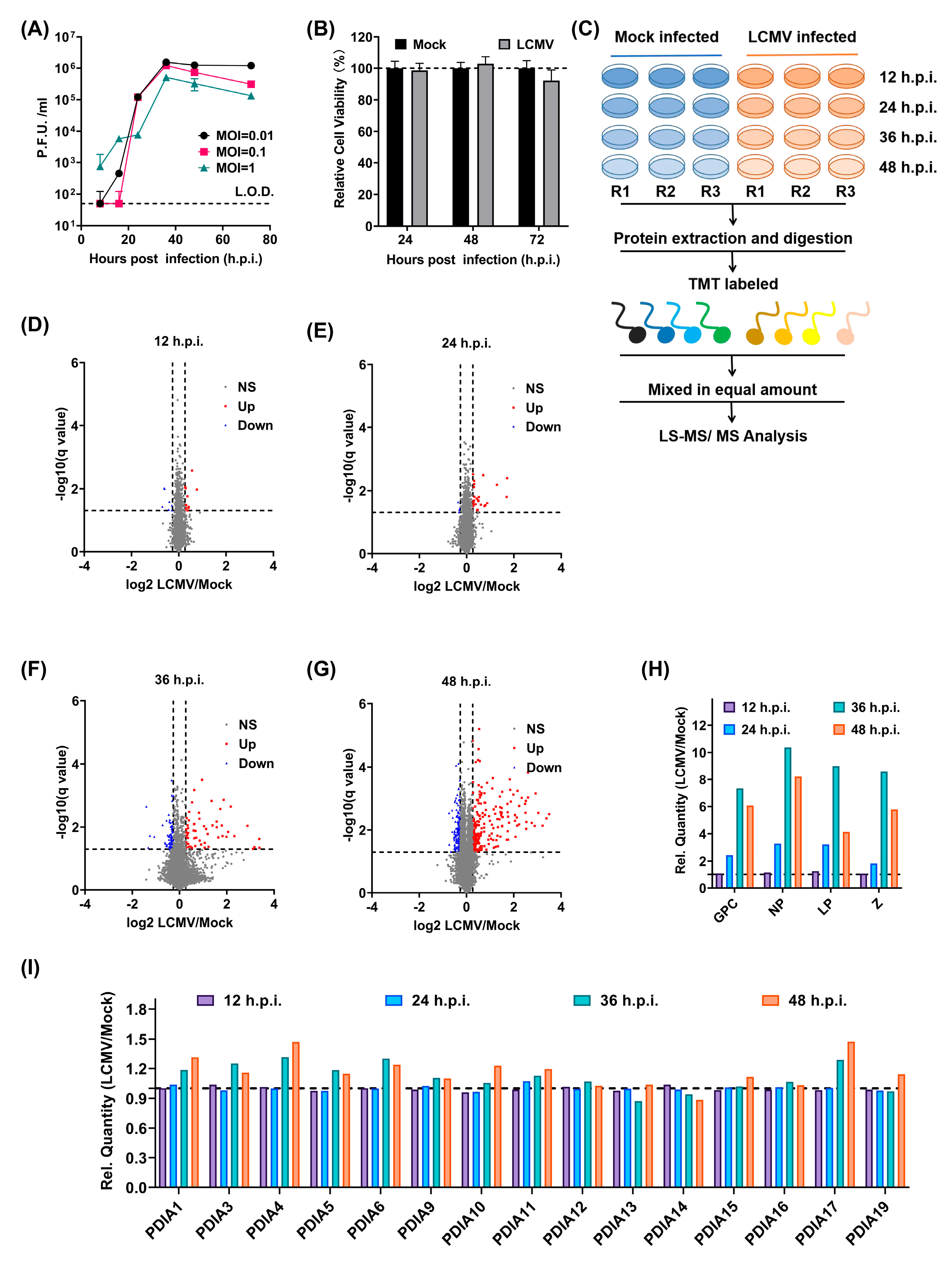
3.1. Identification of New Players in LCMV Infection through a Quantitative Proteomic Screen
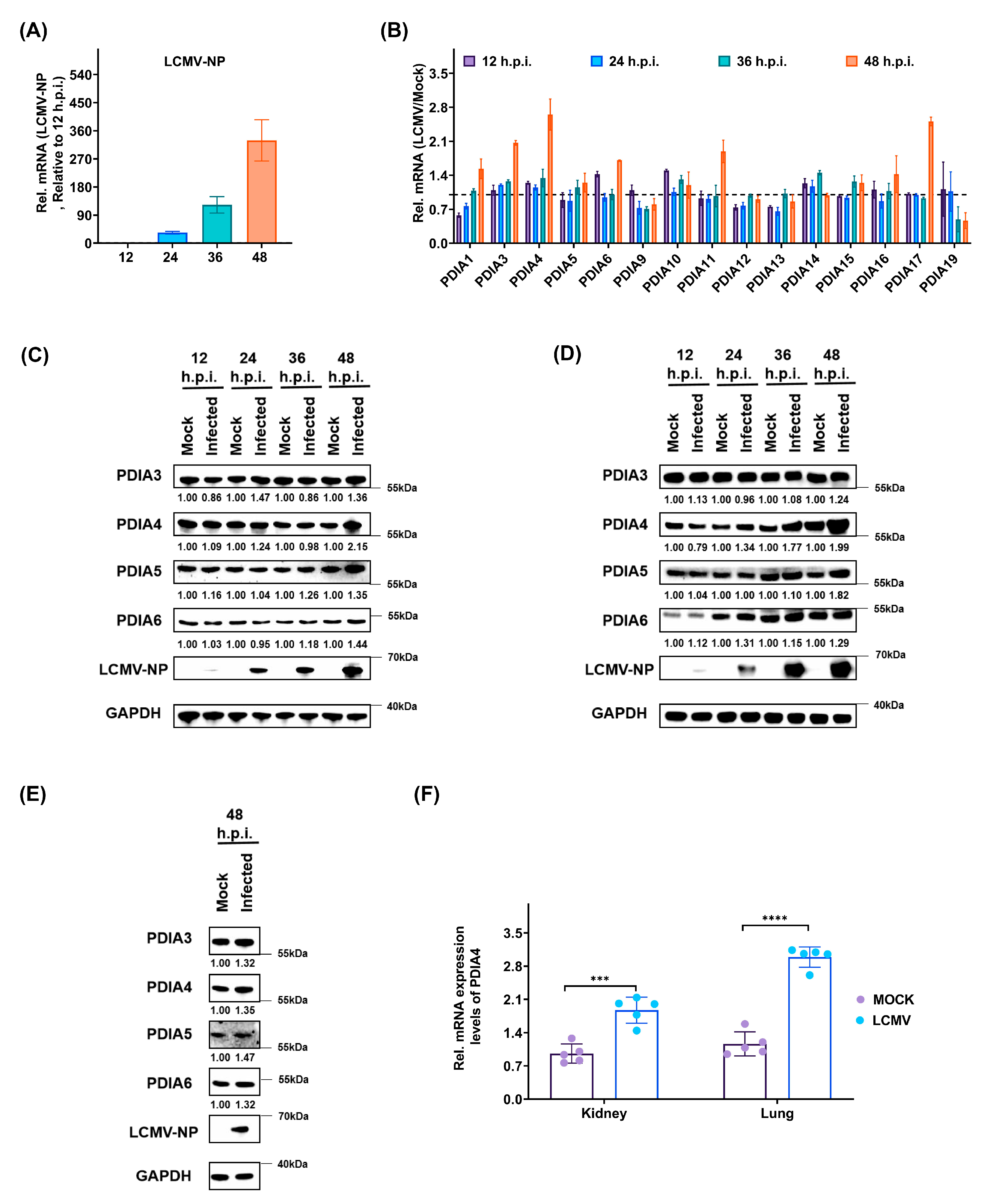
3.2. LCMV Infection Promotes the Expression of PDIs, Especially PDIA4, Both In Vitro and In Vivo
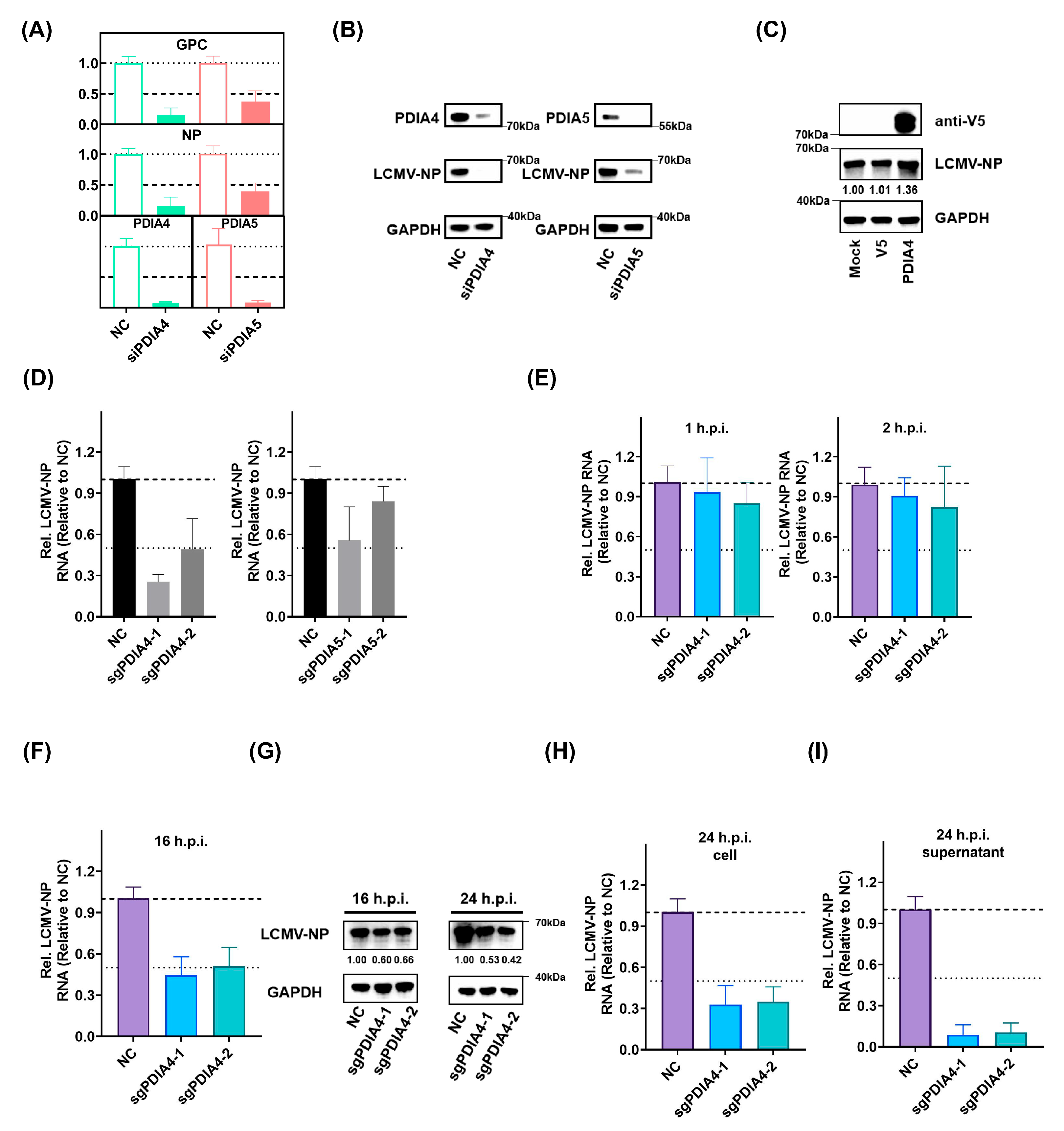
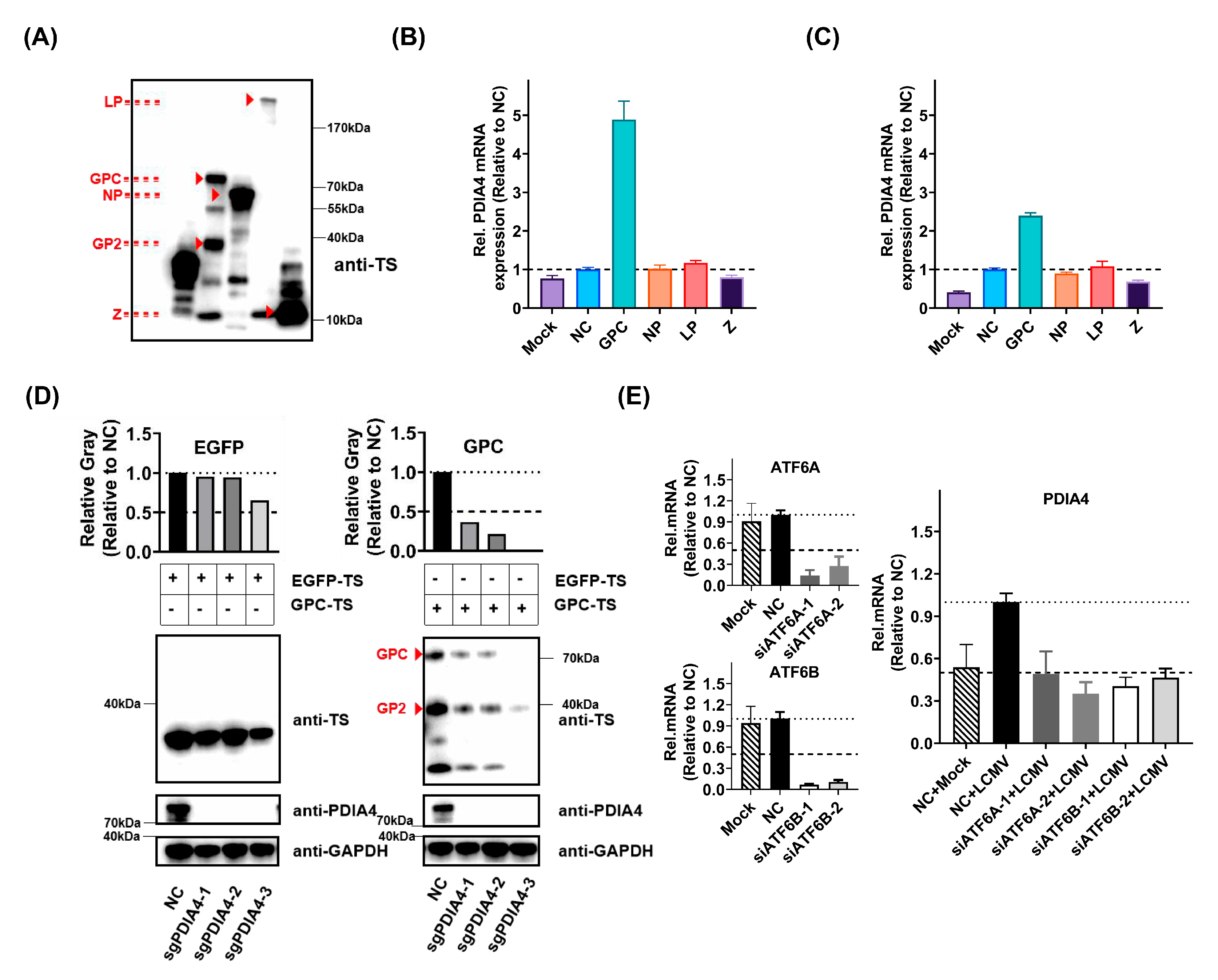
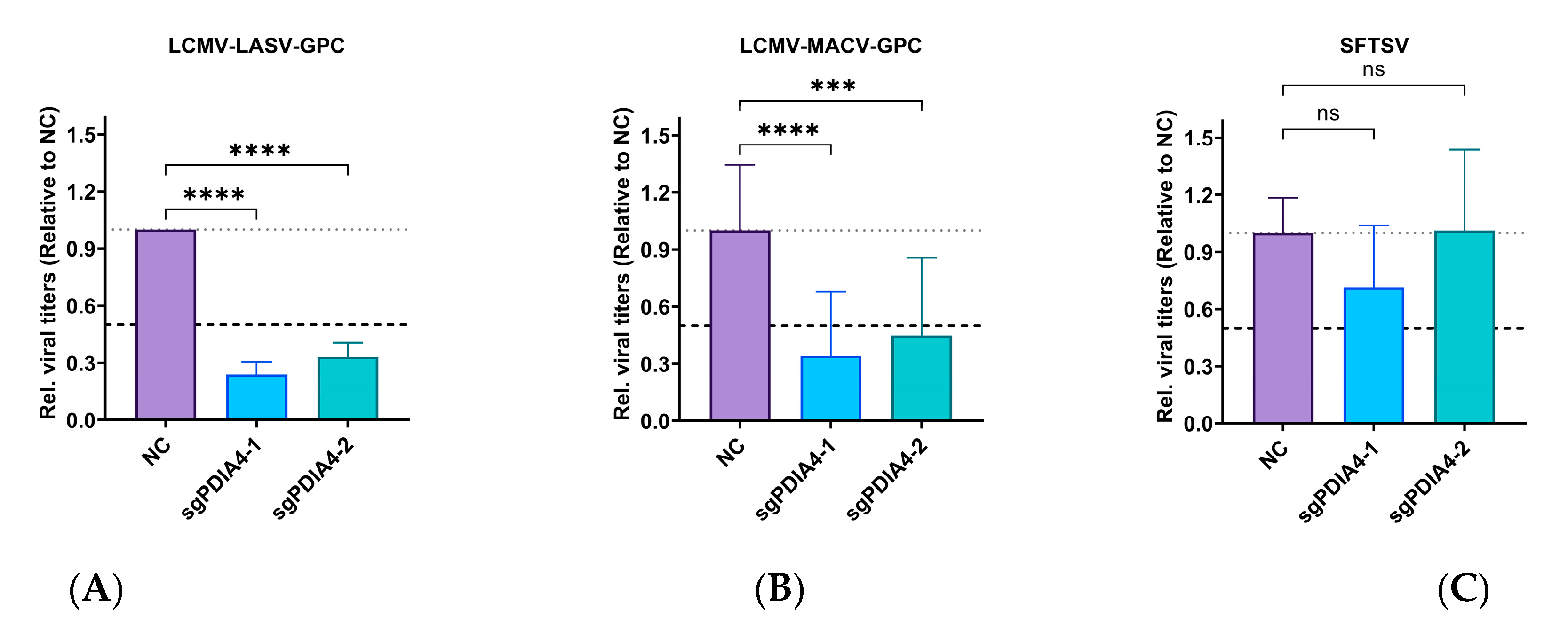
3.3. PDIA4 Is a Positive Regulator in LCMV Infection
3.4. ATF6-Mediated PDIA4 Inhibition Suppresses LCMV Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bentim Góes, L.G.; Fischer, C.; Almeida Campos, A.C.; de Carvalho, C.; Moreira-Soto, A.; Ambar, G.; Ruckert da Rosa, A.; de Oliveira, D.C.; Jo, W.K.; Cruz-Neto, A.P.; et al. Highly Diverse Arenaviruses in Neotropical Bats, Brazil. Emerg. Infect. Dis. 2022, 28, 2528–2533. [Google Scholar] [CrossRef] [PubMed]
- Rivers, T.M.; McNair Scott, T.F. Meningitis in man caused by a filterable virus. Science 1935, 81, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Yun, N.E.; Walker, D.H. Pathogenesis of Lassa fever. Viruses 2012, 4, 2031–2048. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Survey of lymphocytic choriomeningitis virus diagnosis and testing–Connecticut, 2005. MMWR Morb. Mortal. Wkly. Rep. 2006, 55, 398–399. [Google Scholar]
- Martínez-Sobrido, L.; de la Torre, J.C. Reporter-Expressing, Replicating-Competent Recombinant Arenaviruses. Viruses 2016, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Pasqual, G.; Burri, D.J.; Pasquato, A.; de la Torre, J.C.; Kunz, S. Role of the host cell’s unfolded protein response in arenavirus infection. J. Virol. 2011, 85, 1662–1670. [Google Scholar] [CrossRef] [PubMed]
- López, N.; Jácamo, R.; Franze-Fernández, M.T. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 2001, 75, 12241–12251. [Google Scholar] [CrossRef]
- Kozlov, G.; Määttänen, P.; Thomas, D.Y.; Gehring, K. A structural overview of the PDI family of proteins. FEBS J. 2010, 277, 3924–3936. [Google Scholar] [CrossRef]
- Parakh, S.; Atkin, J.D. Novel roles for protein disulphide isomerase in disease states: A double edged sword? Front. Cell Dev. Biol. 2015, 3, 30. [Google Scholar] [CrossRef]
- Marciniak, S.J.; Chambers, J.E.; Ron, D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat. Rev. Drug Discov. 2022, 21, 115–140. [Google Scholar] [CrossRef]
- Nakka, V.P.; Prakash-Babu, P.; Vemuganti, R. Crosstalk Between Endoplasmic Reticulum Stress, Oxidative Stress, and Autophagy: Potential Therapeutic Targets for Acute CNS Injuries. Mol. Neurobiol. 2016, 53, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-K.; Blackwood, E.A.; Azizi, K.; Thuerauf, D.J.; Fahem, A.G.; Hofmann, C.; Kaufman, R.J.; Doroudgar, S.; Glembotski, C.C. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ. Res. 2017, 120, 862–875. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Minder, P.; Caì, Y.; Kuhn, J.H.; Yates, J.R.; Torbett, B.E.; de la Torre, J.C. Interactome analysis of the lymphocytic choriomeningitis virus nucleoprotein in infected cells reveals ATPase Na+/K+ transporting subunit Alpha 1 and prohibitin as host-cell factors involved in the life cycle of mammarenaviruses. PLoS Pathog. 2018, 14, e1006892. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.F.; Hsu, S.W.; Huang, S.H.; Chang, C.L.; Feng, C.S.; Huang, M.G.; Chen, T.Y.; Yang, M.T.; Jiang, S.T.; Wen, T.N.; et al. Pdia4 regulates β-cell pathogenesis in diabetes: Molecular mechanism and targeted therapy. EMBO Mol. Med. 2021, 13, e11668. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Qi, L. Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018, 43, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Chang, K.-O. Protein disulfide isomerases as potential therapeutic targets for influenza A and B viruses. Virus Res. 2018, 247, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, J.; Liu, X.; Chai, Q.; Lu, X.; Yao, X.; Yang, Z.; Sun, L.; Johnson, S.F.; Schwartz, R.C.; et al. Protein disulfide isomerases (PDIs) negatively regulate ebolavirus structural glycoprotein expression in the endoplasmic reticulum (ER) via the autophagy-lysosomal pathway. Autophagy 2022, 18, 2350–2367. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hernández, N.; Meyer, L.; López, S.; DuBois, R.M.; Arias, C.F. Protein Disulfide Isomerase A4 Is Involved in Genome Uncoating during Human Astrovirus Cell Entry. Viruses 2020, 13, 53. [Google Scholar] [CrossRef]
- Kuo, T.F.; Chen, T.Y.; Jiang, S.T.; Chen, K.W.; Chiang, Y.M.; Hsu, Y.J.; Liu, Y.J.; Chen, H.M.; Yokoyama, K.K.; Tsai, K.C.; et al. Protein disulfide isomerase a4 acts as a novel regulator of cancer growth through the procaspase pathway. Oncogene 2017, 36, 5484–5496. [Google Scholar] [CrossRef]
- Tsuji, K.; Kida, Y.; Koshikawa, N.; Yamamoto, S.; Shinozaki, Y.; Watanabe, T.; Lin, J.; Nagase, H.; Takenaga, K. Suppression of non-small-cell lung cancer A549 tumor growth by an mtDNA mutation-targeting pyrrole-imidazole polyamide-triphenylphosphonium and a senolytic drug. Cancer Sci. 2022, 113, 1321–1337. [Google Scholar] [CrossRef]
- Wen, Y.; Xu, H.; Wan, W.; Shang, W.; Jin, R.; Zhou, F.; Mei, H.; Wang, J.; Xiao, G.; Chen, H.; et al. Visualizing lymphocytic choriomeningitis virus infection in cells and living mice. iScience 2022, 25, 105090. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Zhu, S.; Li, S.; Shang, W.; Zhang, R.; Li, H.; Liu, W.; Xiao, G.; Peng, K.; Zhang, L. High-Throughput Screening of an FDA-Approved Drug Library Identifies Inhibitors against Arenaviruses and SARS-CoV-2. ACS Infect. Dis. 2021, 7, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, M.J.G.; Moon-Walker, A.; Herlo, R.; Brusic, V.; Stubbs, S.H.; Hastie, K.M.; Saphire, E.O.; Kirchhausen, T.L.; Whelan, S.P.J. CD164 is a host factor for lymphocytic choriomeningitis virus entry. Proc. Natl. Acad. Sci. USA 2022, 119, e2119676119. [Google Scholar] [CrossRef] [PubMed]
- Fletcher-Etherington, A.; Weekes, M.P. Quantitative Temporal Viromics. Annu. Rev. Virol. 2021, 8, 159–181. [Google Scholar] [CrossRef]
- Di Conza, G.; Ho, P.-C. ER Stress Responses: An Emerging Modulator for Innate Immunity. Cells 2020, 9, 695. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, H.; Cheng, Q. PDIA4: The basic characteristics, functions and its potential connection with cancer. Biomed. Pharmacother. 2020, 122, 109688. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, W.; Liu, Y.; Ma, Z.; Xiang, W.; Wen, Y.; Zhang, D.; Li, Y.; Li, Y.; Li, T.; et al. PDIA4 promotes glioblastoma progression via the PI3K/AKT/m-TOR pathway. Biochem. Biophys. Res. Commun. 2022, 597, 83–90. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Xu, H.; Wan, W.; Jian, X.; Jin, R.; Wang, L.; Wang, J.; Xiao, G.; Zhang, L.; Chen, H.; et al. PDIA4 Is a Host Factor Important for Lymphocytic Choriomeningitis Virus Infection. Viruses 2023, 15, 2343. https://doi.org/10.3390/v15122343
Xu M, Xu H, Wan W, Jian X, Jin R, Wang L, Wang J, Xiao G, Zhang L, Chen H, et al. PDIA4 Is a Host Factor Important for Lymphocytic Choriomeningitis Virus Infection. Viruses. 2023; 15(12):2343. https://doi.org/10.3390/v15122343
Chicago/Turabian StyleXu, Mengwei, Huan Xu, Weiwei Wan, Xiaoqin Jian, Runming Jin, Lin Wang, Jingshi Wang, Gengfu Xiao, Leike Zhang, Hongbo Chen, and et al. 2023. "PDIA4 Is a Host Factor Important for Lymphocytic Choriomeningitis Virus Infection" Viruses 15, no. 12: 2343. https://doi.org/10.3390/v15122343
APA StyleXu, M., Xu, H., Wan, W., Jian, X., Jin, R., Wang, L., Wang, J., Xiao, G., Zhang, L., Chen, H., & Wen, Y. (2023). PDIA4 Is a Host Factor Important for Lymphocytic Choriomeningitis Virus Infection. Viruses, 15(12), 2343. https://doi.org/10.3390/v15122343






