Influenza A, like Omicron SARS-CoV-2, Is Similarly Detected in Saliva or Nasopharyngeal Samples via RT-qPCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and RNA Extraction
2.2. Viruses Molecular Detection
2.3. SARS-CoV 2 Genotyping
2.4. Data Analysis
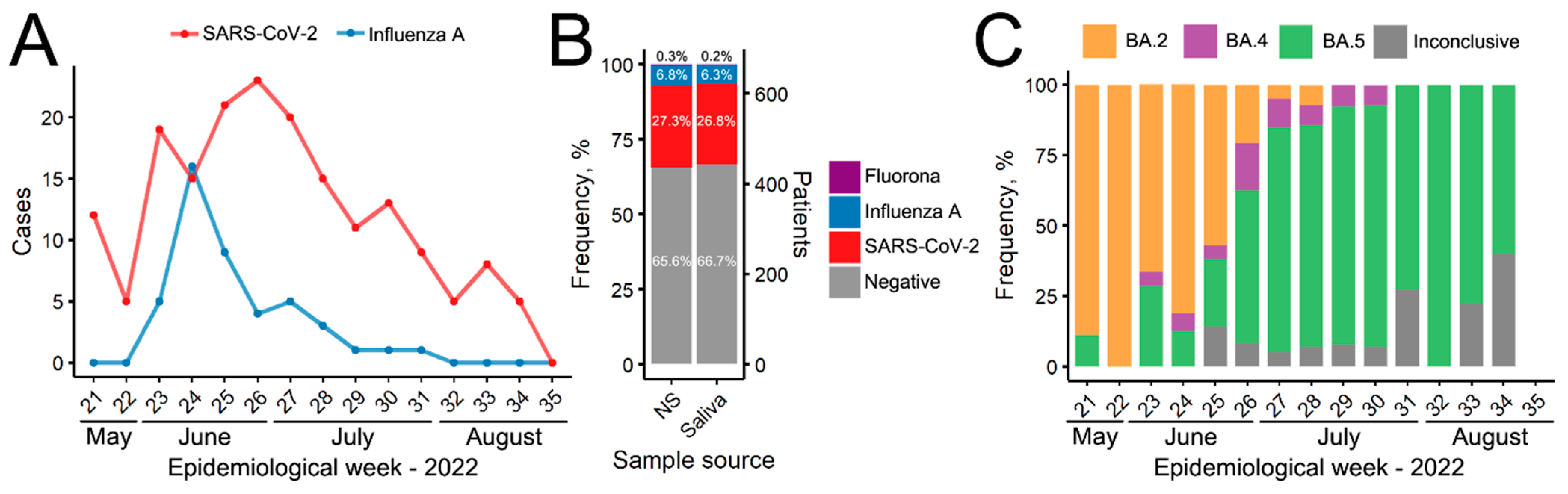
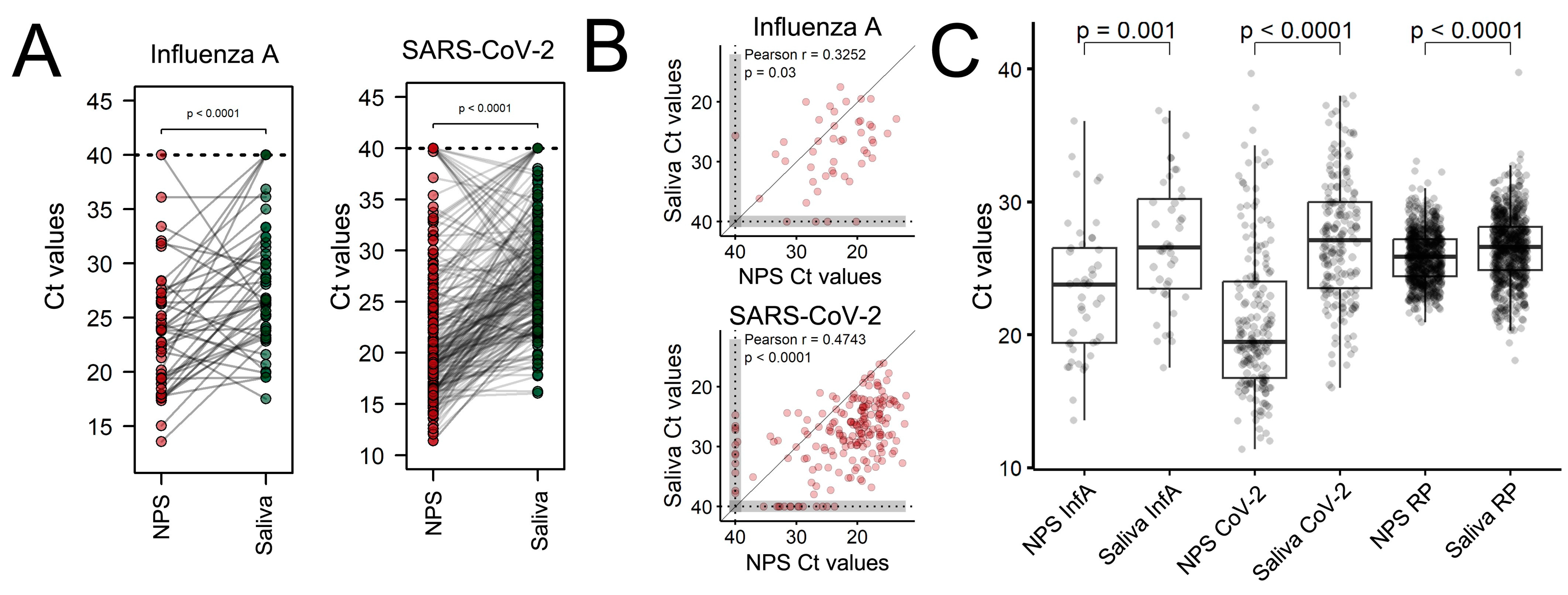
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Keskinocak, P.; Pekgün, P.; Yildirim, I. The balancing role of distribution speed against varying efficacy levels of COVID-19 vaccines under variants. Sci. Rep. 2022, 12, 7493. [Google Scholar] [CrossRef] [PubMed]
- Savela, E.S.; Winnett, A.V.; Romano, A.E.; Porter, M.K.; Shelby, N.; Akana, R.; Ji, J.; Cooper, M.M.; Schlenker, N.W.; Reyes, J.A.; et al. Quantitative SARS-CoV-2 viral-load curves in paired saliva and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. J. Clin. Microbiol. 2022, 60, e01785-21. [Google Scholar] [CrossRef] [PubMed]
- Genelhoud, G.; Adamoski, D.; Spalanzani, R.N.; Bochnia-Bueno, L.; de Oliveira, J.C.; Gradia, D.F.; Bonatto, A.C.; Wassem, R.; Raboni, S.M.; Nogueira, M.B.; et al. Comparison of SARS-CoV-2 molecular detection in nasopharyngeal swab, saliva, and gargle samples. Diagn. Microbiol. Infect. Dis. 2022, 103, 115678. [Google Scholar] [CrossRef] [PubMed]
- Havasi, A.; Visan, S.; Cainap, C.; Cainap, S.S.; Mihaila, A.A.; Pop, L.A. Influenza A, Influenza B, and SARS-CoV-2 similarities and differences—A focus on diagnosis. Front. Microbiol. 2022, 13, 908525. [Google Scholar] [CrossRef] [PubMed]
- Khorramdelazad, H.; Kazemi, M.H.; Najafi, A.; Keykhaee, M.; Zolfaghari Emameh, R.; Falak, R. Immunopathological similarities between COVID-19 and Influenza: Investigating the consequences of coinfection. Microb. Pathog. 2021, 152, 104554. [Google Scholar] [CrossRef]
- Sahajpal, N.S.; Mondal, A.K.; Ananth, S.; Njau, A.; Jones, K.; Ahluwalia, P.; Oza, E.; Ross, T.M.; Kota, V.; Kothandaraman, A.; et al. Clinical validation of a multiplex PCR-based detection assay using saliva or nasopharyngeal samples for SARS-CoV-2, influenza A, and B. Sci. Rep. 2022, 12, 3480. [Google Scholar] [CrossRef]
- ECDC. European Centre for Disease Prevention and Control: Considerations for the Use of Saliva as Sample Material for COVID-19 Testing; ECDC: Stockholm, Sweden, 2021. [Google Scholar]
- CDC. Centers for Disease Control and Prevention. 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel for Emergency Use Only: Instructions for Use: Catalog # 2019-nCoVEUA-01; Division of Viral Diseases: Atlanta, GA, USA, 2019. Available online: https://www.fda.gov/media/134922/download (accessed on 9 October 2023).
- Vogels, C.B.F.; Breban, M.I.; Ott, I.M.; Alpert, T.; Petrone, M.E.; Watkins, A.E.; Kalinich, C.C.; Earnest, R.; Rothman, J.E.; Jesus, J.G.; et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021, 19, e3001236. [Google Scholar] [CrossRef]
- Bilder, L.; Machtei, E.E.; Shenhar, Y.; Kra-Oz, Z.; Basis, F. Salivary detection of H1N1 virus: A clinical feasibility investigation. J. Dent. Res. 2011, 90, 1136–1139. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Yun, S.G.; Kim, M.Y.; Park, K.; Cho, C.H.; Yoon, S.Y.; Nam, M.H.; Lee, C.K.; Cho, Y.-J.; Lim, C.S. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J. Clin. Microbiol. 2017, 55, 115–125. [Google Scholar] [CrossRef]
- Sueki, A.; Matsuda, K.; Yamaguchi, A.; Uehara, M.; Sugano, M.; Uehara, T.; Honda, T. Evaluation of saliva as diagnostic materials for influenza virus infection by PCR-based assays. Clin. Chim. Acta 2016, 453, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Galar, A.; Catalán, P.; Vesperinas, L.; Miguens, I.; Muñoz, I.; García-Espona, A.; Sevillano, J.A.; Andueza, J.A.; Bouza, E.; Muñoz, P. Use of saliva swab for detection of Influenza virus in patients admitted to an emergency department. Microbiol. Spectr. 2021, 9, e0033621. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Yun, S.G.; Nam, J.; Choi, S.-H.; Lim, C.S. The use of saliva specimens for detection of influenza A and B viruses by rapid influenza diagnostic tests. J. Virol. Method. 2017, 243, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Lukas, H.; Tu, J.; Xu, C.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter 2020, 3, 1981–1998. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Pan, Y.; Sun, Z.; Li, J.; Yi, Y.; Yang, J.; Li, G. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens. Bioelectron. 2021, 186, 113309. [Google Scholar] [CrossRef]
- Byrne, R.L.; Kay, G.A.; Kontogianni, K.; Aljayyoussi, G.; Brown, L.; Collins, A.M.; Cuevas, L.E.; Ferreira, D.M.; Fraser, A.J.; Garrod, G.; et al. Saliva alternative to upper respiratory swabs for SARS-CoV-2 diagnosis. Emerg. Infect. Dis. 2020, 26, 2770–2771. [Google Scholar] [CrossRef]
- Walker, N.F.; Byrne, R.L.; Howard, A.; Nikolaou, E.; Farrar, M.; Glynn, S.; Cheliotis, K.S.; Atienzar, A.I.C.; Davies, K.; Reiné, J.; et al. Detection of SARS-CoV-2 infection by saliva and nasopharyngeal sampling in frontline healthcare workers: An observational cohort study. PLoS ONE 2023, 18, e0280908. [Google Scholar] [CrossRef]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 2020, 370, m2516. [Google Scholar] [CrossRef]
- Law, S.; Tovar, M.A.; Franke, M.F.; Calderon, R.; Palomino, S.; Valderrama, G.; Llanos, F.; Velásquez, G.E.; Mitnick, C.D.; Lecca, L. Low detection rate of RT-PCR-confirmed COVID-19 using IgM/IgG rapid antibody tests in a large community sample in Lima, Peru. BMC Infect. Dis. 2023, 23, 62. [Google Scholar] [CrossRef]
- Kritikos, A.; Caruana, G.; Lazor-Blanchet, C.; Currat, M.; Chiche, J.D.; Vollenweider, P.; Bart, P.A.; Opota, O.; Greub, G. Comparison of nasopharyngeal and saliva swab nucleic acid amplification and rapid antigen testing to detect Omicron SARS-CoV-2 Variant of Concern: A prospective clinical trial (OMICRON). Microbiol. Spectr. 2022, 10, e0392322. [Google Scholar] [CrossRef]
- Caixeta, D.C.; Paranhos, L.R.; Blumenberg, C.; Garcia-Júnior, M.A.; Guevara-Vega, M.; Taveira, E.B.; Nunes, M.A.C.; Cunha, T.M.; Jardim, A.C.G.; Flores-Mir, C.; et al. Salivary SARS-CoV-2 RNA for diagnosis of COVID-19 patients: A systematic review and meta-analysis of diagnostic accuracy. Jpn. Dent. Sci. Rev. 2023, 59, 219–238. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimi, N.; Delaunay-Moisan, A.; Hill, C.; Le Teuff, G.; Rupprecht, J.-F.; Thuret, J.-Y.; Chaltiel, D.; Potier, M.-C. Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis. PLoS ONE 2021, 16, e0253007. [Google Scholar] [CrossRef] [PubMed]
- Moreira, V.M.; Mascarenhas, P.; Machado, V.; Botelho, J.; Mendes, J.J.; Taveira, N.; Almeida, M.G. Diagnosis of SARS-Cov-2 infection by RT-PCR using specimens other than naso- and oropharyngeal swabs: A systematic review and meta-analysis. Diagnostics 2021, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Basurrah, M.A.; Han, J.H.; Kim, S.W.; Hwang, S.H. The diagnostic accuracy of RT-PCR from self-collected saliva versus nasopharyngeal sampling: A systematic review and meta-analysis. Saudi Med. J. 2022, 43, 9–30. [Google Scholar] [CrossRef]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef]
- Okoturo, E.; Amure, M. SARS-CoV-2 saliva testing using RT-PCR: A systematic review. Int. J. Infect. Dis. 2022, 121, 166–171. [Google Scholar] [CrossRef]
- Migueres, M.; Mansuy, J.-M.; Vasseur, S.; Claverie, N.; Lougarre, C.; Soulier, F.; Trémeaux, P.; Izopet, J. Omicron Wave SARS-CoV-2 Diagnosis: Evaluation of saliva, anterior nasal, and nasopharyngeal swab samples. Microbiol. Spectr. 2022, 10, e0252122. [Google Scholar] [CrossRef]
- Cornette, M.; Decaesteker, B.; Martens, G.A.; Vandecandelaere, P.; Jonckheere, S. From Delta to Omicron SARS-CoV-2 variant: Switch to saliva sampling for higher detection rate. J. Clin. Virol. Plus 2022, 2, 100090. [Google Scholar] [CrossRef]
- Uršič, T.; Kogoj, R.; Šikonja, J.; Roškarič, D.; Jevšnik Virant, M.; Bogovič, P.; Petrovec, M. Performance of nasopharyngeal swab and saliva in detecting Delta and Omicron SARS-CoV-2 variants. J. Med. Virol. 2022, 94, 4704–4711. [Google Scholar] [CrossRef]
- Ahti, J.; Österback, R.; Keskitalo, A.; Mokkala, K.; Vidbäck, S.; Veikkolainen, V.; Vuorinen, T.; Peltola, V.; Hakanen, A.J.; Waris, M.; et al. Diagnostic performance and tolerability of saliva and nasopharyngeal swab specimens in the detection of SARS-CoV-2 by RT-PCR. Microbiol. Spectr. 2023, 11, e0532422. [Google Scholar] [CrossRef]
- Bordi, L.; Sberna, G.; Lalle, E.; Fabeni, L.; Mazzotta, V.; Lanini, S.; Corpolongo, A.; Garbuglia, A.R.; Nicastri, E.; Girardi, E.; et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva samples from patients infected with Omicron variant. Int. J. Mol. Sci. 2023, 24, 4847. [Google Scholar] [CrossRef] [PubMed]
- Gusev, E.; Sarapultsev, A.; Solomatina, L.; Chereshnev, V. SARS-CoV-2-Specific immune response and the pathogenesis of COVID-19. Int. J. Mol. Sci. 2022, 23, 1716. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Li, Y.; Yu, B.M.N.; Brinkman, T.; Wong, D.T. Characterization of RNA in Saliva. Clin. Chem. 2006, 52, 988–994. [Google Scholar] [CrossRef]
- Ott, I.M.; Strine, M.S.; Watkins, A.E.; Boot, M.; Kalinich, C.C.; Harden, C.A.; Vogels, C.B.F.; Casanovas-Massana, A.; Moore, A.J.; Muenker, M.C.; et al. Stability of SARS-CoV-2 RNA in Nonsupplemented Saliva. Emerg Infect Dis. 2021, 27, 1146–1150. [Google Scholar] [CrossRef]
- Bru, S.; Brotons, P.; Jordan, I.; Alsina, L.; Henares, D.; Carballar, R.; Sevilla, M.F.; Barrabeig, I.; Fumado, V.; Baro, B.; et al. Association between soluble angiotensin-converting enzyme 2 in saliva and SARS-CoV-2 infection: A cross-sectional study. Sci. Rep. 2023, 13, 5985. [Google Scholar] [CrossRef]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates. Virus Genes 2022, 58, 255–269. [Google Scholar] [CrossRef]
- Stevens, L.; de Buyl, S.; Mognetti, B.M. The sliding motility of the bacilliform virions of Influenza A viruses. Soft Matter 2023, 19, 4491–4501. [Google Scholar] [CrossRef]
- Kai-Feng, H.; Yi-Chen, S.; Bing-Hong, C.; Jeng-Fan, L.; Chao-Min, C.; Cho-Yi, C.; Cheng-Hsien, W.; Shou-Yen, K. New COVID-19 saliva-based test: How good is it compared with the current nasopharyngeal or throat swab test? J. Chin. Med. Assoc. 2020, 83, 891–894. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abreu, H.; Royer, C.A.; Poitevin, C.G.; Kohler, A.F.; Rodrigues, A.C.; Raboni, S.M.; Nogueira, M.B.; Cardoso, P.H.; Arruda, M.B.; Baptista, P.A.d.S.; et al. Influenza A, like Omicron SARS-CoV-2, Is Similarly Detected in Saliva or Nasopharyngeal Samples via RT-qPCR. Viruses 2023, 15, 2352. https://doi.org/10.3390/v15122352
Abreu H, Royer CA, Poitevin CG, Kohler AF, Rodrigues AC, Raboni SM, Nogueira MB, Cardoso PH, Arruda MB, Baptista PAdS, et al. Influenza A, like Omicron SARS-CoV-2, Is Similarly Detected in Saliva or Nasopharyngeal Samples via RT-qPCR. Viruses. 2023; 15(12):2352. https://doi.org/10.3390/v15122352
Chicago/Turabian StyleAbreu, Hellen, Carla Adriane Royer, Carolina Gracia Poitevin, Ana Flávia Kohler, Ana Carolina Rodrigues, Sonia Mara Raboni, Meri Bordignon Nogueira, Pedro Henrique Cardoso, Monica Barcellos Arruda, Patrícia Alvarez da Silva Baptista, and et al. 2023. "Influenza A, like Omicron SARS-CoV-2, Is Similarly Detected in Saliva or Nasopharyngeal Samples via RT-qPCR" Viruses 15, no. 12: 2352. https://doi.org/10.3390/v15122352
APA StyleAbreu, H., Royer, C. A., Poitevin, C. G., Kohler, A. F., Rodrigues, A. C., Raboni, S. M., Nogueira, M. B., Cardoso, P. H., Arruda, M. B., Baptista, P. A. d. S., Bonatto, A. C., Gradia, D. F., Adamoski, D., Maltempi de Souza, E., & Carvalho de Oliveira, J. (2023). Influenza A, like Omicron SARS-CoV-2, Is Similarly Detected in Saliva or Nasopharyngeal Samples via RT-qPCR. Viruses, 15(12), 2352. https://doi.org/10.3390/v15122352







