Genetic Diversity, Evolutionary Dynamics, and Ongoing Spread of Pedilanthus Leaf Curl Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Multiple Sequence Alignment
2.2. Time and Tree Root Estimation
2.3. Phylogeny and Phylogeography
2.4. PeLCV Population Structure Assay
2.5. Network Analysis
2.6. Estimation of Nucleotide Substitution Rate
2.7. Selection Pressure Analysis
2.8. Recombination Analysis
3. Results
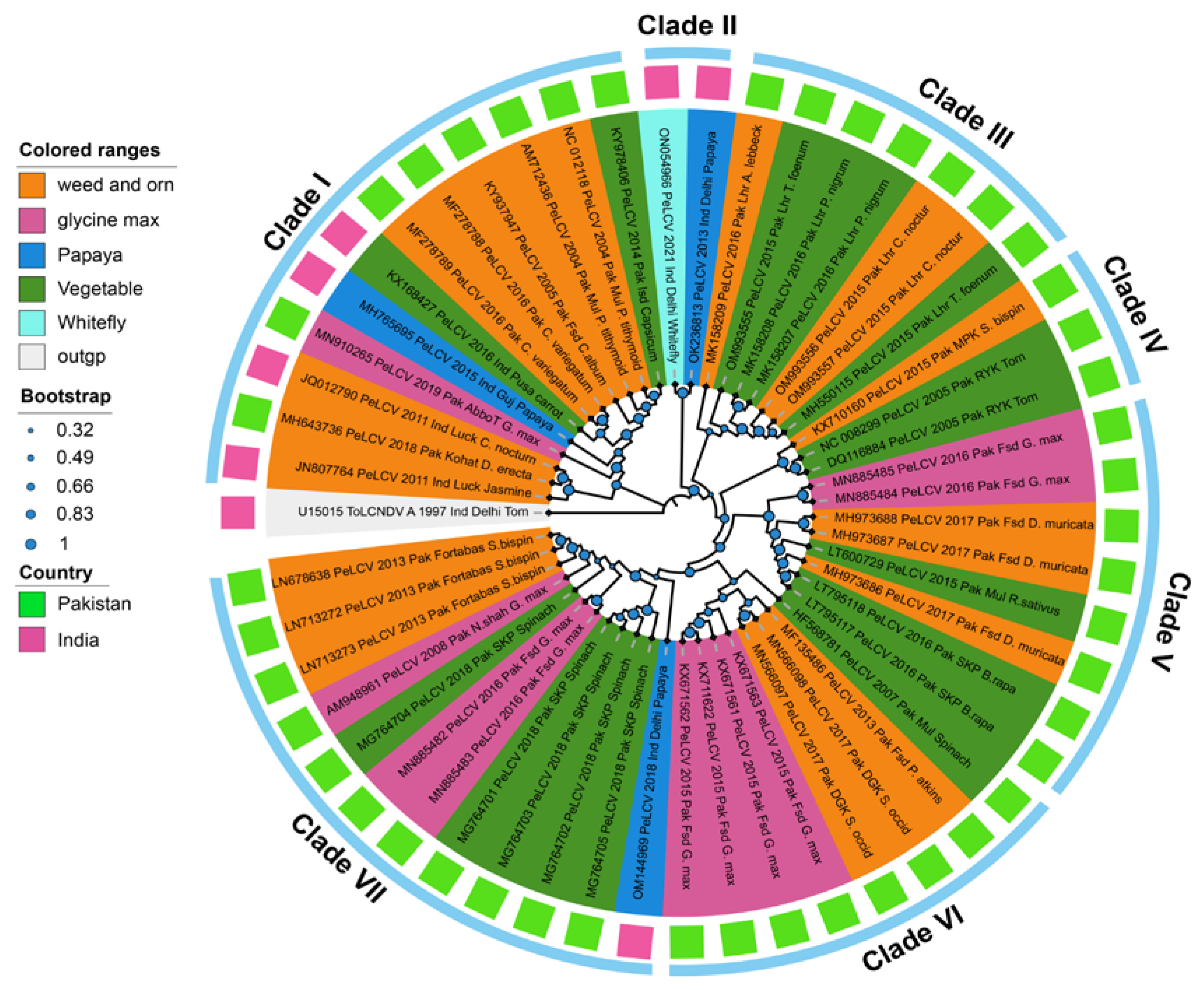
3.1. Evolutionary Time Estimation, Phylogeny, and Network Analysis
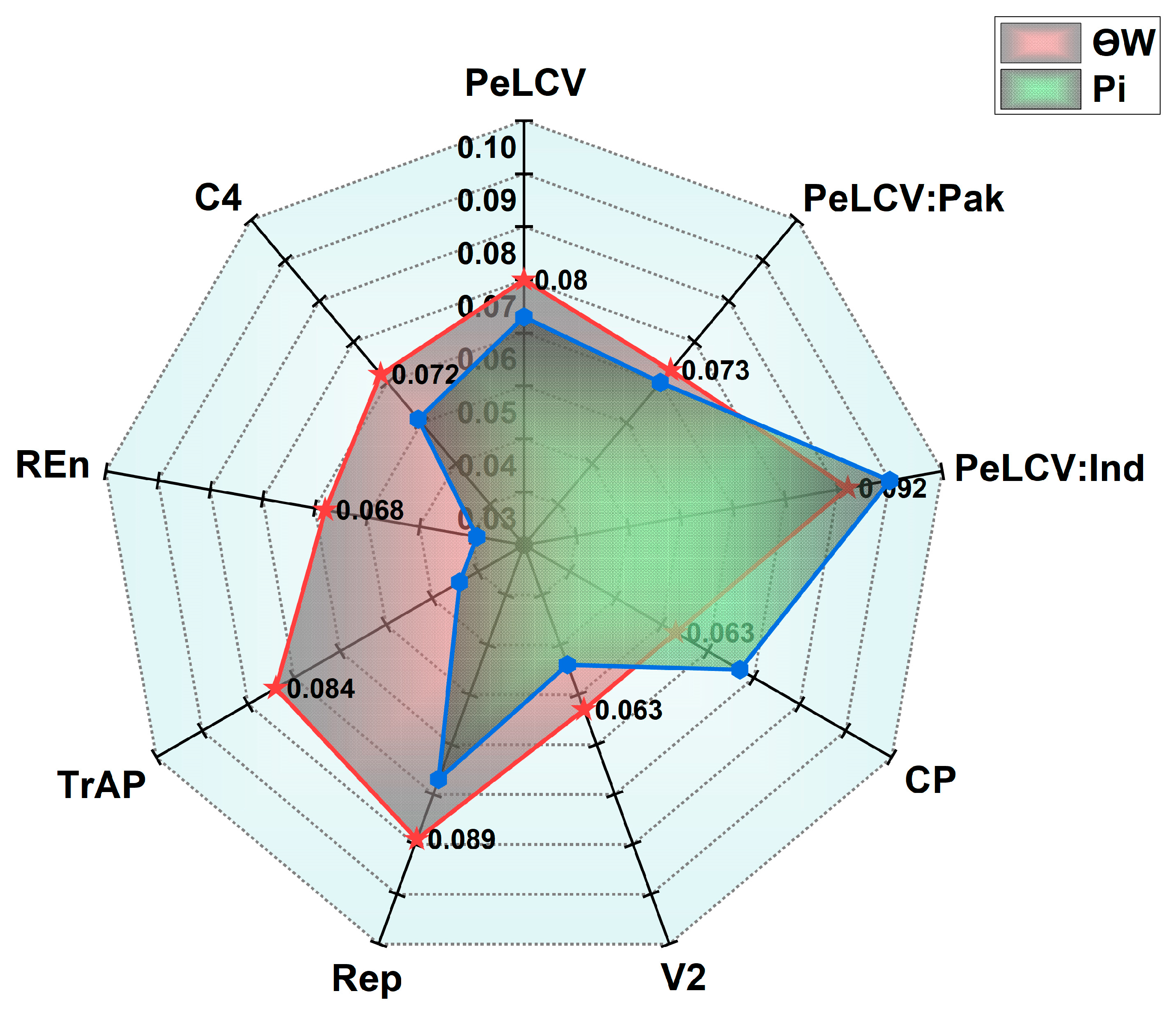
3.2. Genetic Diversity Indices
3.3. Neutrality Indices
3.4. Estimation of Selection Pressure
3.5. Evolutionary Rate Estimation
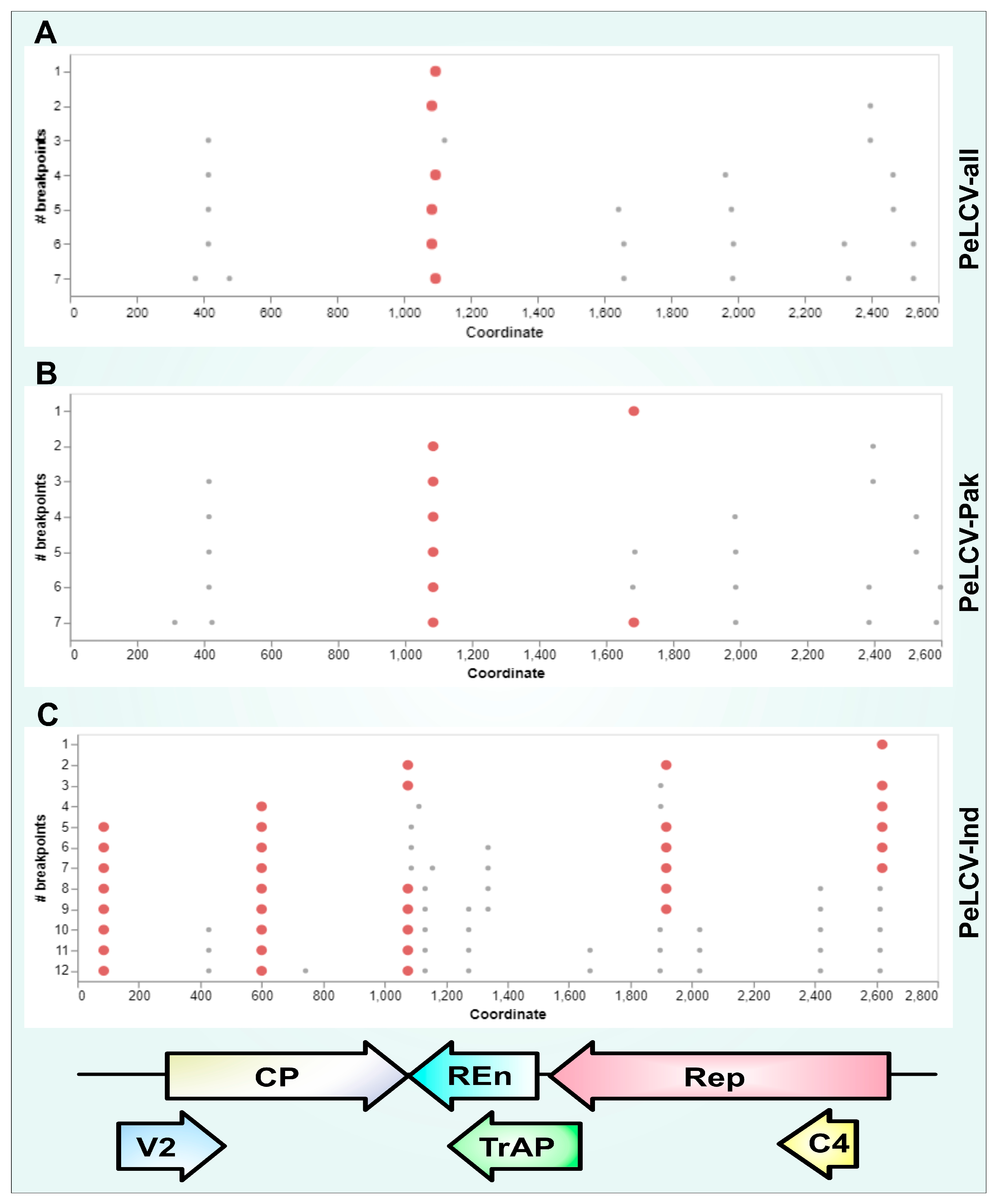
3.6. Recombination Analysis
3.7. Time-Scaled Phylogeography of PeLCV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, W.M.O. The Whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) Interaction with Geminivirus-Infected Host Plants: Bemisia tabaci, Host Plants and Geminiviruses, 1st ed.; Springer: Dordrecht, The Netherlands, 2011. [Google Scholar]
- Fiallo-Olivé, E.; Pan, L.L.; Liu, S.S.; Navas-Castillo, J. Transmission of begomoviruses and other whitefly-borne viruses: Dependence on the vector species. Phytopathology 2020, 110, 10–17. [Google Scholar] [CrossRef]
- Masood, M.; Amin, I.; Hassan, I.; Mansoor, S.; Brown, J.K.; Briddon, R.W. Diversity and ddistribution of cryptic species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex in Pakistan. J. Econ. Entomol. 2017, 110, 2295–2300. [Google Scholar] [CrossRef]
- Naga, K.C.; Siddappa, S.; Kumar, R.; Tiwari, R.K.; Subhash, S.; Verma, G.; Buckseth, T.; Bairwa, A.; Sharma, S.; Katare, S.; et al. A new record of Asia II 5 genetic group of Bemisia tabaci (Gennadius) in the major potato growing areas of India and its relationship with tomato leaf curl New Delhi virus infecting potato. 3 Biotech 2021, 11, 421. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World Management of Geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef]
- Tahir, M.; Haider, S.; Iqbal, J.; Briddon, R.W. Association of a distinct begomovirus and a betasatellite with leaf curl symptoms in Pedilanthus tithymaloides. J. Phytopath. 2009, 157, 188–193. [Google Scholar] [CrossRef]
- Saritha, R.K.; Shrawan, S.; Kalia, P.; Jain, R.K.; Baranwal, V.K. Association of Pedilanthus leaf curl virus and satellites with carrot (Daucus carota) in India. Plant Dis. 2017, 101, 843. [Google Scholar] [CrossRef]
- Ijaz, S.; Mubin, M.; Rehman, M.S.N.U. Molecular analysis of Pedilanthus leaf curl virus and associated satellites infecting Chenopodium album in pakistan. Pak. J. Agric. Sci. 2020, 57, 425–432. [Google Scholar]
- Yasmin, S.; Raja, N.I.; Hameed, S.; Brown, J.K. First association of Pedilanthus leaf curl virus, Papaya leaf curl virus, Cotton leaf curl Kokhran virus, and Papaya leaf curl betasatellite with symptomatic chilli pepper in Pakistan. Plant Dis. 2017, 101, 2155. [Google Scholar] [CrossRef]
- Shakir, S.; Nawaz-Ul-Rehman, M.S.; Mubin, M.; Ali, Z. Characterization, phylogeny and recombination analysis of Pedilanthus leaf curl virus-Petunia isolate and its associated betasatellite. Virol. J. 2018, 15, 134. [Google Scholar] [CrossRef]
- Ismail, H.; Hassan, I.; Zubair, M.; Khan, Z.; Sarfaraz, S.; Jamil, N.; Mansoor, S.; Asad, S.; Amin, I. First report of Pedilanthus leaf curl virus, Tobacco leaf curl betasatellite and Guar leaf curl alphasatellite infecting radish (Raphanus sativus) in Pakistan. Plant Dis. 2017, 101, 845. [Google Scholar] [CrossRef]
- Ilyas, M.; Qazi, J.; Mansoor, S.; Briddon, R.W. Genetic diversity and phylogeography of begomoviruses infecting legumes in Pakistan. J. Gen. Virol. 2010, 91, 2091–2101. [Google Scholar] [CrossRef]
- Zaidi, S.S.A.; Amin, I.; Iqbal, Z.; Pervaiz Akhtar, K.; Scheffler, B.E.; Mansoor, S. Sesbania bispinosa, a new host of a begomovirus-betasatellite complex in Pakistan. Can. J. Plant Pathol. 2016, 38, 107–111. [Google Scholar] [CrossRef]
- Tahir, M.N.; Hameed, A.; Amin, I.; Mansoor, S. Characterization of a begomovirus-betasatellite complex, producing defective molecules in spinach (Spinacia oleracea L.), a new host for begomovirus and betasatellite complex in Pakistan. Plant Pathol. J. 2017, 33, 514–521. [Google Scholar] [CrossRef]
- Munir, S.; Khurshid, M.; Kanwal, H.; Hussain, M.; Sattar, M.N.; Ali, I.; Rehman, A.-u.; Iqbal, Z. Identification of Pedilanthus leaf curl virus and associated betasatellite infecting turnip in Pakistan. J. Plant Pathol. 2018, 100, 317–321. [Google Scholar] [CrossRef]
- Sattar, M.N.; Qurashi, F.; Iqbal, Z.; El-Beltagi, H.S.; Khurshid, M. Occurrence and molecular characteristics of Pedilanthus leaf curl virus complex from the new hosts fenugreek and night jessamine in Pakistan. Physiol. Mol. Plant Pathol. 2023, 126, 102045. [Google Scholar] [CrossRef]
- Kumar, J.; Singh, S.; Kumar, A.; Khan, J.; Tuli, R. Detection and characterization of a new betasatellite: Variation in disease symptoms of tomato leaf curl Pakistan virus-India due to associated betasatellite. Arch. Virol. 2013, 158, 257–261. [Google Scholar] [CrossRef]
- Yousaf, S.; Rasool, G.; Akhtar, S. First report of Pedilanthus leaf curl virus and Ageratum conyzoides symptomless alphasatellite infecting Cassia occidentalis in Pakistan. J. Plant Pathol. 2021, 103, 731–732. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef]
- Iqbal, Z.; Shafiq, M.; Mansoor, S.; Briddon, R.W. Analysis of the effects of the mutation of selected genes of Pedilanthus leaf curl virus on infectivity, symptoms and the maintenance of tobacco leaf curl betasatellite. Can. J. Plant Pathol. 2022, 44, 767–779. [Google Scholar] [CrossRef]
- Roossinck, M.J. Mechansims of plant virus evolution. Annu. Rev. Phytopathol. 1997, 35, 191–209. [Google Scholar] [CrossRef]
- Seal, S.E.; Jeger, M.J.; Van den Bosch, F.; Maramorosch, K.; Shatkin, A.J.; Thresh, J.M. Begomovirus evolution and disease management. Adv. Virus Res. 2006, 67, 297–316. [Google Scholar]
- Simmonds, P.; Aiewsakun, P.; Katzourakis, A. Prisoners of war—Host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 2019, 17, 321–328. [Google Scholar] [CrossRef]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef]
- Nigam, D. Genomic variation and diversification in begomovirus genome in implication to host and vector adaptation. Plants 2021, 10, 1706. [Google Scholar] [CrossRef]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef]
- Duffy, S.; Holmes, E.C. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus Tomato yellow leaf curl virus (TYLCV). J. Virol. 2008, 82, 957–965. [Google Scholar] [CrossRef]
- Lima, A.T.M.; Silva, J.C.F.; Silva, F.N.; Castillo-Urquiza, G.P.; Silva, F.F.; Seah, Y.M.; Mizubuti, E.S.G.; Duffy, S.; Zerbini, F.M. The diversification of begomovirus populations is predominantly driven by mutational dynamics. Virus Evol. 2017, 3, vex005. [Google Scholar] [CrossRef]
- Sattar, M.N.; Khurshid, M.; El-Beltagi, H.S.; Iqbal, Z. Identification and estimation of sequence variation dynamics of Tomato leaf curl Palampur virus and betasatellite complex infecting a new weed host. Biotechnol. Biotechnol. Equip. 2022, 36, 609–619. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bedford, I.D.; Tsai, J.H.; Markham, P.G. Analysis of the nucleotide sequence of the treehopper-transmitted geminivirus, tomato pseudo-curly top virus, suggests a recombinant origin. Virology 1996, 219, 387–394. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Pita, J.S.; Fondong, V.N.; Sangre, A.; Otim-Nape, G.W.; Ogwal, S.; Fauquet, C.M. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 2001, 82, 655–665. [Google Scholar] [CrossRef]
- Sanz, A.I.; Fraile, A.; García-Arenal, F.; Zhou, X.; Robinson, D.J.; Khalid, S.; Butt, T.; Harrison, B.D. Multiple infection, recombination and genome relationships among begomovirus isolates found in cotton and other plants in Pakistan. J. Gen. Virol. 2000, 81, 1839–1849. [Google Scholar] [CrossRef]
- García-Andrés, S.; Accotto, G.P.; Navas-Castillo, J.; Moriones, E. Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the tomato yellow leaf curl disease in the Mediterranean basin. Virology 2007, 359, 302–312. [Google Scholar] [CrossRef]
- Varsani, A.; Shepherd, D.N.; Monjane, A.L.; Owor, B.E.; Erdmann, J.B.; Rybicki, E.P.; Peterschmitt, M.; Briddon, R.W.; Markham, P.G.; Oluwafemi, S.; et al. Recombination, decreased host specificity and increased mobility may have driven the emergence of maize streak virus as an agricultural pathogen. J. Gen. Virol. 2008, 89, 2063–2074. [Google Scholar] [CrossRef]
- Xavier, C.A.D.; Godinho, M.T.; Mar, T.B.; Ferro, C.G.; Sande, O.F.L.; Silva, J.C.; Ramos-Sobrinho, R.; Nascimento, R.N.; Assunção, I.; Lima, G.S.A.; et al. Evolutionary dynamics of bipartite begomoviruses revealed by complete genome analysis. Mol. Ecol. 2021, 30, 3747–3767. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2006, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Bielejec, F.; Baele, G.; Vrancken, B.; Suchard, M.A.; Rambaut, A.; Lemey, P. SpreaD3: Interactive visualization of spatiotemporal history and trait evolutionary processes. Mol. Biol. Evol. 2016, 33, 2167–2169. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Sattar, N.M.; Iqbal, Z.; Najabat Ali, S.; Amin, I.; Shafiq, M.; Khurshid, M. Natural occurrence of mesta yellow vein mosaic virus and DNA-satellites in ornamental sunflower (Helianthus spp.) in Pakistan. Saudi J. Biol. Sci. 2021, 28, 6621–6630. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Wisler, G.C.; Norris, R.F. Interactions between weeds and cultivated plants as related to management of plant pathogens. Weed Sci. 2005, 53, 914–917. [Google Scholar] [CrossRef]
- Polston, J.E.; Barro, P.J.; Boykin, L.M. Transmission specificities of plant viruses with the newly identified species of the Bemisia tabaci species complex. Pest Manag. Sci. 2014, 70, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.L.; Cui, X.Y.; Chen, Q.F.; Wang, X.W.; Liu, S.S. Cotton Leaf Curl Disease: Which Whitefly Is the Vector? Phytopathol. 2018, 108, 1172–1183. [Google Scholar] [CrossRef]
- Masood, M.; Briddon, R.W. Transmission of cotton leaf curl disease: Answer to a long-standing question. Virus Genes 2018, 54, 743–745. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Verma, R.K.; Pandey, V.; Srivastava, A.; Sharma, P.; Gaur, R.; Ali, A. Role of diversity and recombination in the emergence of chilli leaf curl virus. Pathogens 2022, 11, 529. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Pandey, V.; Sahu, A.K.; Yadav, D.; Al-Sadi, A.M.; Shahid, M.S.; Gaur, R.K. Evolutionary dynamics of begomoviruses and its satellites infecting papaya in India. Front. Microbiol. 2022, 13, 879413. [Google Scholar] [CrossRef]
- Mar, T.B.; Xavier, C.A.D.; Lima, A.T.M.; Nogueira, A.M.; Silva, J.C.F.; Ramos-Sobrinho, R.; Lau, D.; Zerbini, F.M. Genetic variability and population structure of the New World begomovirus Euphorbia yellow mosaic virus. J. Gen. Virol. 2017, 98, 1537–1551. [Google Scholar] [CrossRef]
- Sobrinho, R.R.; Xavier, C.A.D.; Pereira, H.M.d.B.; Lima, G.S.d.A.; Assunção, I.P.; Mizubuti, E.S.G.; Duffy, S.; Zerbini, F.M. Contrasting genetic structure between two begomoviruses infecting the same leguminous hosts. J. Gen. Virol. 2014, 95, 2540–2552. [Google Scholar] [CrossRef]
- Mishra, M.; Verma, R.K.; Marwal, A.; Sharma, P.; Gaur, R.K. Biology and interaction of the natural occurrence of distinct monopartite begomoviruses associated with satellites in Capsicum annum from India. Front. Microbiol. 2020, 11, 512957. [Google Scholar] [CrossRef]
- Farooq, T.; Umar, M.; She, X.; Tang, Y.; He, Z. Molecular phylogenetics and evolutionary analysis of a highly recombinant begomovirus, Cotton leaf curl Multan virus, and associated satellites. Virus Evol. 2021, 7, veab054. [Google Scholar] [CrossRef]
- Garcia-Arenal, F.; Fraile, A.; Malpica, J.M. Variability and genetic structure of plant virus populations. Ann. Rev. Phytopathol. 2001, 39, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Holmes, E.C. Validation of high rates of nucleotide substitution in geminiviruses: Phylogenetic evidence from East African cassava mosaic viruses. J. Gen. Virol. 2009, 90, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S. The spread of Tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef] [PubMed]
- Lefeuvre, P.; Lett, J.-M.; Varsani, A.; Martin, D.P. Widely conserved recombination patterns amongst single stranded DNA viruses. J. Virol. 2009, 83, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.E.; van den Bosch, F.; Jeger, M.J. Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. Crit. Rev. Plant Sci. 2006, 25, 23–46. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Moriones, E. Recombination as a motor of host switches and virus emergence: Geminiviruses as case studies. Curr. Opin. Virol. 2015, 10, 14–19. [Google Scholar] [CrossRef]
- Prasanna, H.C.; Rai, M. Detection and frequency of recombination in tomato-infecting begomoviruses of South and Southeast Asia. Virol. J. 2007, 4, 111. [Google Scholar] [CrossRef]
- Martin, D.P.; Lefeuvre, P.; Varsani, A.; Hoareau, M.; Semegni, J.-Y.; Dijoux, B.; Vincent, C.; Reynaud, B.; Lett, J.-M. Complex recombination patterns arising during geminivirus coinfections preserve and demarcate biologically important intra-genome interaction networks. PLoS Pathog. 2011, 7, e1002203. [Google Scholar] [CrossRef]

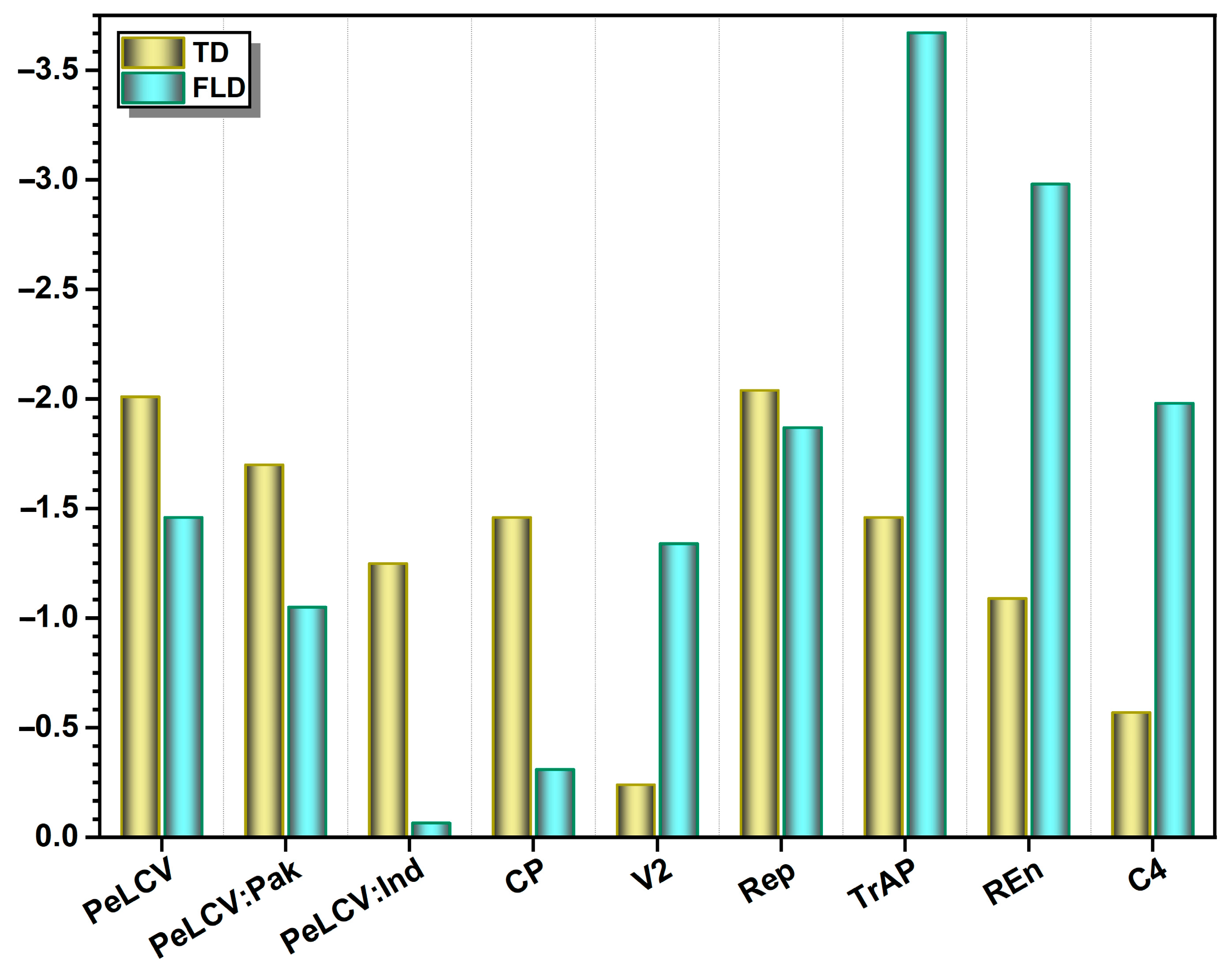

| Dataset | No. of Seq | InDel Sites | S | Eta (h) | K | Neutrality Test | |
|---|---|---|---|---|---|---|---|
| TD | FLD | ||||||
| PeLCV all | 52 | 86 | 851 | 1057 | 172.08 | −2.01 | −1.46 |
| PeLCV-Pak | 45 | 152 | 794 | 949 | 175.01 | −1.70 | −1.05 |
| PeLCV-Ind | 07 | 62 | 609 | 684 | 272.0 | −1.25 | −0.066 |
| CP | 52 | 2 | 219 | 266 | 59.56 | −1.46 | −0.31 |
| V2 | 52 | 10 | 92 | 115 | 18.38 | −0.24 | −1.34 |
| Rep | 52 | 18 | 351 | 433 | 67.41 | −2.04 | −1.87 |
| TrAP | 52 | 24 | 146 | 182 | 16.93 | −1.46 | −3.67 |
| REn | 52 | 21 | 118 | 145 | 14.97 | −1.09 | −2.98 |
| C4 | 52 | 0 | 99 | 118 | 18.77 | −0.57 | −1.98 |
| ORFs | Best Model | Mean Distance (d) | dN | dS | dN/dS | FUBAR Posterior Probability (p ≤ 0.9) | SLAC (p ≤ 0.05) | ||
|---|---|---|---|---|---|---|---|---|---|
| PS | NS | PS | NS | ||||||
| V2 | K2 + G | 0.058 ± 0.007 | 0.034 ± 0.007 | 0.150 ± 0.028 | 4.86 | 8 | 110 | 0 | 8 |
| CP | T92 + G | 0.088 ± 0.007 | 0.049 ± 0.006 | 0.214 ± 0.025 | 0.23 | 2 | 99 | 0 | 50 |
| Rep | HKY + G | 0.086 ± 0.008 | 0.083 ± 0.006 | 0.061 ± 0.011 | 1.36 | 3 | 112 | 0 | 46 |
| TrAP | K2 + G | 0.048 ± 0.006 | 0.035 ± 0.006 | 0.087 ± 0.016 | 0.40 | 3 | 11 | 1 | 10 |
| REn | T92 + G | 0.042 ± 0.006 | 0.028 ± 0.006 | 0.090 ± 0.018 | 0.31 | 4 | 11 | 0 | 8 |
| C4 | K2 + G | 0.066 ± 0.009 | 0.044 ± 0.003 | 0.07 ± 0.010 | 0.63 | 9 | 4 | 0 | 3 |
| Dataset | PeLCV-All | PeLCV-Pak | PeLCV-Ind | |||
|---|---|---|---|---|---|---|
| Strict Clock | Relaxed Clock | Strict Clock | Relaxed Clock | Strict Clock | Relaxed Clock | |
| Mean nt substitution rate (site−1 year−1) | 8.22 × 10−4 (225) | 8.15 × 10−4 (216) | 1.44 × 10−4 (863) | 1.46 × 10−4 (539) | 8.20 × 10−4 (673) | 8.13 × 10−4 (523) |
| At 95% HPD interval | 8.27 × 10−4, 8.19 × 10−4 | 8.16 × 10−4, 8.13 × 10−4 | 1.44 × 10−4, 1.43 × 10−4 | 1.45 × 10−4, 1.44 × 10−4 | 8.26 × 10−4, 8.16 × 10−4 | 8.14 × 10−4, 8.11 × 10−4 |
| ORFs | Clock Type | Mean nt Substitution Rate (Site−1 Year−1) | At 95% HPD Interval | CoP1 Mu | CoP2 Mu | CoP3 Mu |
|---|---|---|---|---|---|---|
| V2 | Strict | 2.39 × 10−2 (215) | 2.76 × 10−2, 2.09 × 10−2 | 1.308 (415) | 0.404 (213) | 1.289 (520) |
| Relaxed | 2.48 × 10−2 (288) | 2.61 × 10−3, 2.42 × 10−3 | 1.311 (1899) | 0.332 (4381) | 1.357 (1488) | |
| CP | Strict | 8.03 × 10−3 (1508) | 9.01 × 10−3, 3.44 × 10−3 | 0.397 (4210) | 1.086 (6754) | 1.52 (7591) |
| Relaxed | 1.97 × 10−4 (1451) | 1.76 × 10−4, 2.06 × 10−4 | 0.466 (650) | 0.928 (541) | 1.608 (519) | |
| Rep | Strict | 7.1 × 10−3 (901) | 7.51 × 10−3, 6.87 × 10−3 | 0.504 (7237) | 1.483 (7734) | 1.013 (5767) |
| Relaxed | 7.21 × 10−3 (831) | 7.25 × 10−3, 7.16 × 10−4 | 0.495 (7669) | 1.486 (7998) | 1.019 (6349) | |
| TrAP | Strict | 2.73 × 10−4 (820) | 3.71 × 10−4, 3.21 × 10−4 | 1.058 (7597) | 0.776 (8213) | 1.166 (8536) |
| Relaxed | 2.72 × 10−4 (715) | 2.79 × 10−4, 2.65 × 10−4 | 1.071 (1334) | 0.739 (4361) | 1.180 (364) | |
| REn | Strict | 2.52 × 10−4 (712) | 2.97 × 10−4, 2.17 × 10−4 | 1.085 (7149) | 0.612 (6959) | 1.302 (7699) |
| Relaxed | 2.49 × 10−4 (661) | 2.57 × 10−4, 2.43 × 10−4 | 0.978 (7294) | 0.636 (7486) | 1.386 (6969) | |
| C4 | Strict | 1.81 × 10−3 (881) | 2.08 × 10−3, 1.72 × 10−3 | 0.271 (7795) | 2.105 (6322) | 0.624 (6826) |
| Relaxed | 2.13 × 10−3 (505) | 2.17 × 10−3, 2.09 × 10−3 | 0.282 (2573) | 2.093 (2611) | 0.625 (2573) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, Z.; Shafiq, M.; Sattar, M.N.; Ali, I.; Khurshid, M.; Farooq, U.; Munir, M. Genetic Diversity, Evolutionary Dynamics, and Ongoing Spread of Pedilanthus Leaf Curl Virus. Viruses 2023, 15, 2358. https://doi.org/10.3390/v15122358
Iqbal Z, Shafiq M, Sattar MN, Ali I, Khurshid M, Farooq U, Munir M. Genetic Diversity, Evolutionary Dynamics, and Ongoing Spread of Pedilanthus Leaf Curl Virus. Viruses. 2023; 15(12):2358. https://doi.org/10.3390/v15122358
Chicago/Turabian StyleIqbal, Zafar, Muhammad Shafiq, Muhammad Naeem Sattar, Irfan Ali, Muhammad Khurshid, Umer Farooq, and Muhammad Munir. 2023. "Genetic Diversity, Evolutionary Dynamics, and Ongoing Spread of Pedilanthus Leaf Curl Virus" Viruses 15, no. 12: 2358. https://doi.org/10.3390/v15122358
APA StyleIqbal, Z., Shafiq, M., Sattar, M. N., Ali, I., Khurshid, M., Farooq, U., & Munir, M. (2023). Genetic Diversity, Evolutionary Dynamics, and Ongoing Spread of Pedilanthus Leaf Curl Virus. Viruses, 15(12), 2358. https://doi.org/10.3390/v15122358






