The Novel Porcine Parvoviruses: Current State of Knowledge and Their Possible Implications in Clinical Syndromes in Pigs
Abstract
1. Introduction
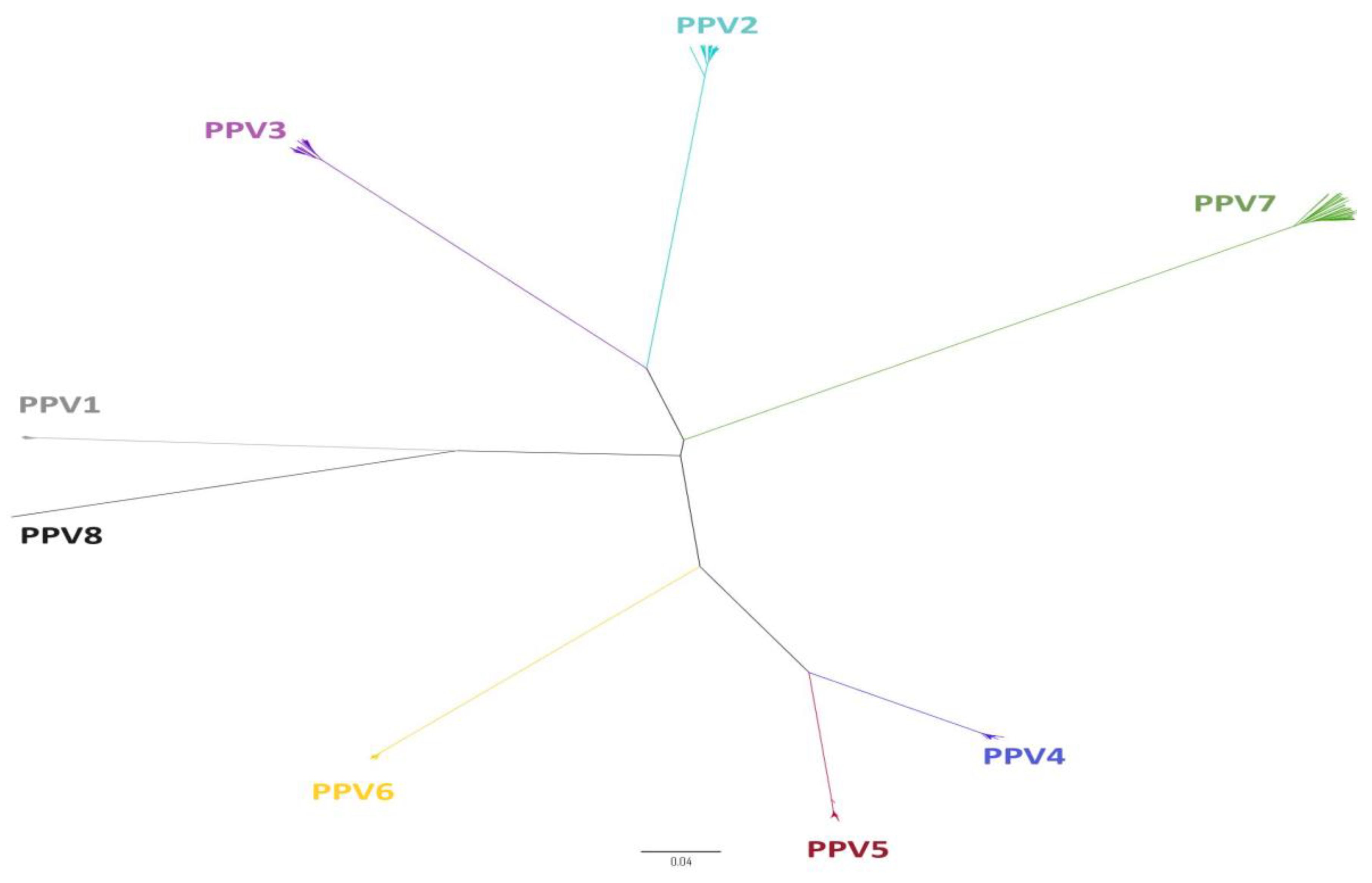
2. Classification and Molecular Organization of Porcine Parvovirus
| Virus (GenBank Accession Number) | ICTV Current Classification (2022) | Genome Size (nt ¶) | ORF 1 (aa #) | ORF 2 (aa) | Year of the Discovery | Evolutionary Rates (Substitutions/Site/Year) * | Reference |
|---|---|---|---|---|---|---|---|
| PPV1 (L23427) | Protoparvovirus Ungulate 1 | 5075 | 662 | 729 | 1965 | 6.22 × 10−5 | [36] |
| PPV2 (KM926355) | Tetraparvovirus Ungulate 3 | 5444 | 662 | 1032 | 2001 | 1.35 × 10−4 | [37] |
| PPV3 (EU200677) | Tetraparvovirus Ungulate 2 | 5114 | 637 | 926 | 2008 | 8.16 × 10−4 | [10] |
| PPV4 (FJ872544) | Copiparvovirus Ungulate 2 | 5905 | 588 | 728 | 2010 | 4.70 × 10−4 | [33] |
| PPV5 (JX896318) | Not yet classified | 5805 | 601 | 991 | 2013 | 6.95 × 10−5 | [38] |
| PPV6 (KY094494) | Copiparvovirus Ungulate 4 | 6148 | 662 | 1189 | 2014 | 4.90 × 10−4 | [12] |
| PPV7 (MG902949) | Chaphamaparvovirus Ungulate 1 | 4103 | 672 | 469 | 2015 | 4.85 × 10−3 | [14] |
| PPV8 (OP021638) | Not yet classified | 4380 | 601 | 701 | 2022 | Not reported | [15] |
3. Evolution of Porcine Parvoviruses
3.1. Mutations in Porcine Parvovirus
3.2. Recombination of Porcine Parvoviruses
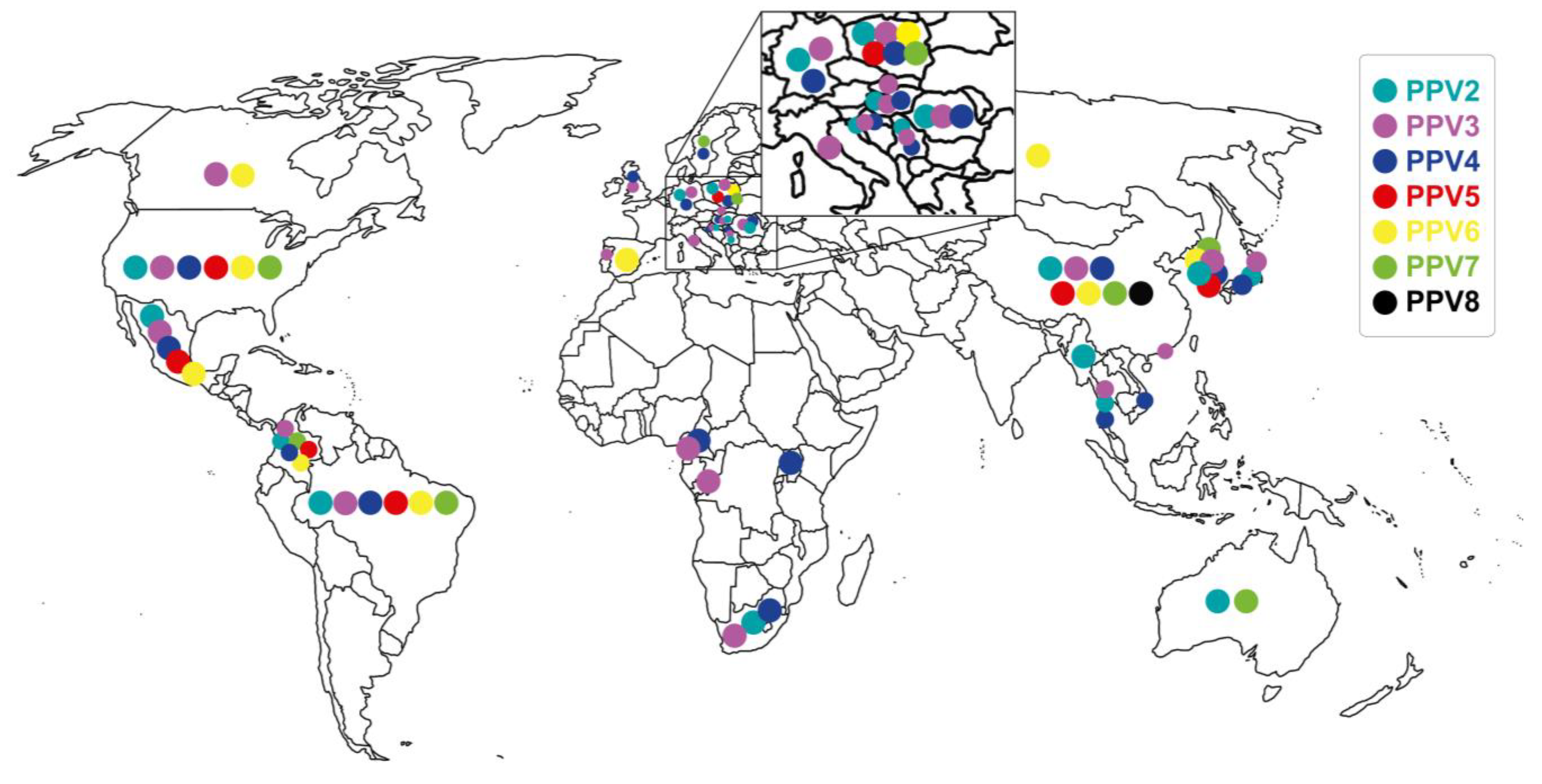
4. Epidemiology of Porcine Parvoviruses
4.1. Distribution of Porcine Parvoviruses
4.2. Prevalence by Age Groups of Novel Porcine Parvovirus
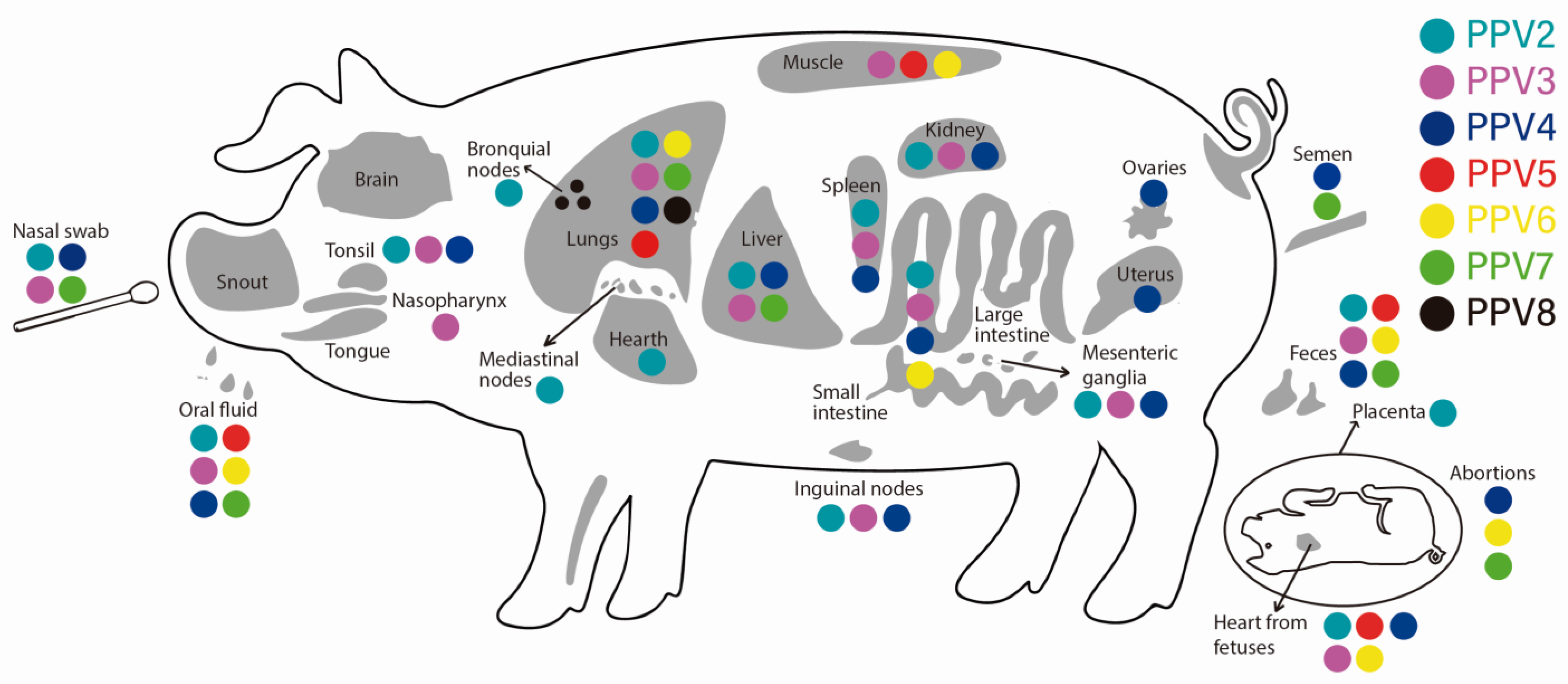
5. Tropism of Porcine Parvoviruses
6. Association of Porcine Parvoviruses with Disease
6.1. Porcine Reproductive Failure (PRF)
6.2. Porcine Respiratory Disease Complex (PRDC)
| Detection or Prevalence (%) in Different Types of Samples | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | L | F | NS | OF | OP | H | T | Sp | LN | K | Li | ABF | Sm | PFET | Country | Reference | |
| PPV2 | 5.4 | 21–51 | 6 | Hungary | [17,73] | ||||||||||||
| 25 | Romania | [25] | |||||||||||||||
| 55 | 78 | Germany | [74] | ||||||||||||||
| 19–54 | 19.4 | 48.7 | Poland | [80,95,119] | |||||||||||||
| 6 | 30 | 2 | 28 | 43 | 29–52 | 100 | 25 | 2.8 | Europe | [24] | |||||||
| 11–12 | 2.5–86.6 | 7.5–12.7 | 2.5 | Republic of Korea | [60,72,98] | ||||||||||||
| 4–8.7 | 73 | 0.33 | 22.5–39.5 | 22.5 | China | [22,58,71,97] | |||||||||||
| 58–100 | Japan | [57] | |||||||||||||||
| 22 | 32 | 56 | Vietnam | [120] | |||||||||||||
| 83 | Thailand | [121] | |||||||||||||||
| 35–55 | 20.7–42.7 | 7.6 | 7 | USA | [11,13,18,122] | ||||||||||||
| 90 | Mexico | [78] | |||||||||||||||
| 21.8 | South Africa | [75] | |||||||||||||||
| PPV3 | 35–48 | 53.8 | 10.8 | 10 | 22–45 | 19.5 | 46–72 | 71–79 | 53 | China | [10,22,58,82,97] | ||||||
| 73 | Thailand | [121] | |||||||||||||||
| 16.8 | 18.4 | Vietnam | [120] | ||||||||||||||
| 3.8–4.2 | 7.5–17.5 | 8.4–10 | Republic of Korea | [60,98] | |||||||||||||
| 20 | Germany | [74] | |||||||||||||||
| 14.4 | 2.1 | 5.6 | 9 | Hungary | [73] | ||||||||||||
| 17.5 | Romania | [25] | |||||||||||||||
| 7.7–15.4 | 5.6 | 26.7 | Poland | [80,96,119] | |||||||||||||
| 6 | 6 | 6 | 3 | 3–17 | 100 | Europe | [24] | ||||||||||
| 21.8 | South Africa | [75] | |||||||||||||||
| 17.5 | DR Congo | [123] | |||||||||||||||
| 9.4 | 9.1 | USA | [13] | ||||||||||||||
| 65 | 68.5 | 57.5 | 57.5 | 54 | Brazil | [85] | |||||||||||
| 59 | Mexico | [78] | |||||||||||||||
| PPV4 | 5.41 | 3.85 | 4.3–51.3 | China | [22,58,97] | ||||||||||||
| 3.8–9.5 | 8.5 | 2.5–6 | Republic of Korea | [60,98] | |||||||||||||
| 44 | Thailand | [121] | |||||||||||||||
| 7.6 | Vietnam | [120] | |||||||||||||||
| 2.7 | 2.3 | 7 | 50 | 50 | Hungary | [73] | |||||||||||
| 17.5 | Romania | [25] | |||||||||||||||
| 7 | Germany | [74] | |||||||||||||||
| 2.4–10 | 13.9 | 28.7 | Poland | [80,95,119] | |||||||||||||
| 3 | 8 | 2 | 7 | 4–7 | 15 | 0.7 | Europe | [24] | |||||||||
| 5.9 | 4.1–7.5 | 4 | USA | [11,13] | |||||||||||||
| 25.9 | Mexico | [78] | |||||||||||||||
| 84 | 26.6 | 18.2 | 38.5 | Brazil | [90,124] | ||||||||||||
| 43.6 | South Africa | [75] | |||||||||||||||
| PPV5 | 3.4 | 3–6.6 | 5.4 | USA | [11,13] | ||||||||||||
| 8.2 | 34.6 | 9.19 | China | [22,97] | |||||||||||||
| 3.8–9.5 | 21.6 | 5.6 | Republic of Korea | [60,98] | |||||||||||||
| 4–19 | 21 | 41.3 | Poland | [80,95,119] | |||||||||||||
| 32.4 | Mexico | [78] | |||||||||||||||
| PPV6 | 3.8–75 | 34.6 | 12.5 | 3.9 | 50 | China | [12,22,97] | ||||||||||
| 12.5–19.4 | 2.5–27.7 | 2.5–12.4 | 2.5 | Republic of Korea | [60,98] | ||||||||||||
| 6–25.8 | 17.1 | 38 | Poland | [80,95,119] | |||||||||||||
| 9.4 | Rusia | [125] | |||||||||||||||
| 13.2 | USA | [93] | |||||||||||||||
| 74.7 | Mexico | [78] | |||||||||||||||
| PPV7 | 2.1 | 13.8 | 17.5 | 17.2 | USA | [14] | |||||||||||
| 24–65 | 28.3 | 19.8 | 17.4 | 15.4 | 55 | China | [63,96,117,126] | ||||||||||
| 1.9–35 | 7.5–74 | 3.6–22.5 | 24 | 1.5 | Republic of Korea | [39,98,100] | |||||||||||
| 15–19.6 | 39 | Poland | [23,119] | ||||||||||||||
| 8.6 | Brazil | [99] | |||||||||||||||
| 21.4 | 6 | Colombia | [62] | ||||||||||||||
6.3. Association of Porcine Parvovirus with Other Pathologies
6.4. Viral Coinfections between Porcine Parvovirus and Other Viruses
| Novel Porcine Parvovirus | Coinfections | Reference |
|---|---|---|
| PPV2 | PCV2 | [13,24,60,73,78,97,119,122] |
| PRRSV | [18,60,97] | |
| PPV1 to PPV7 | [25,75,78,98] | |
| PRV | [97] | |
| SIV | [18] | |
| PTTV1 | [97] | |
| PBo-like V, PBoV | [75] | |
| M. hyopneumoniae | [18] | |
| PPV3 | PCV2 | [13,24,73,78,82,97,119] |
| PRRSV | [60,97] | |
| PPV1 to PPV7 | [75,78,98] | |
| PRV | [97] | |
| PTTV1 | [97] | |
| PBo-like V, PBoV | [75] | |
| SIV | [83] | |
| M. hyopneumoniae | [83] | |
| PPV4 | PCV2 | [13,78,88,97,119] |
| PRRSV | [60] | |
| PPV1 to PPV7 | [75,78,98] | |
| PBo-like V, PBoV | [75] | |
| PTTV1, PTTV2 | [88,97] | |
| PPV5 | PCV2 | [13,78,97,119] |
| PRRSV | [60,97] | |
| PRV | [97] | |
| PTTV1 | [97] | |
| PPV1 to PPV7 | [78,98] | |
| PPV6 | PCV2 | [13,78,97,119] |
| PRRSV | [60,93,97] | |
| PPRV | [97] | |
| PPV1 to PPV5 and PPV7 | [78,98] | |
| PPV7 | PCV2 | [63,96,119] |
| PRRSV | [60,63] | |
| PCV3 | [63,117] | |
| PEDV | [62] | |
| PPV2 to PPV6 | [98] | |
| PPV8 | PRRSV | [15] |
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pénzes, J.J.; Söderlund-Venermo, M.; Canuti, M.; Eis-Hübinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.-M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. Ictv Report Consortium ICTV virus taxonomy profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.C.; Tomlinson, J.E.; Lopez-Astacio, R.A.; Parrish, C.R.; Van de Walle, G.R. Small but mighty: Old and new parvoviruses of veterinary significance. Virol. J. 2021, 18, 210. [Google Scholar] [CrossRef] [PubMed]
- Mengeling, W.L.; Lager, K.M.; Vorwald, A.C. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim. Reprod. Sci. 2000, 60–61, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, E.M. Update on canine parvoviral enteritis. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 1307–1325. [Google Scholar] [CrossRef] [PubMed]
- Stuetzer, B.; Hartmann, K. Feline parvovirus infection and associated diseases. Vet. J. 2014, 201, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Higgins, P.G.; Davis, L.R.; Willman, J.S.; Jones, S.E.; Kidd, I.M.; Pattison, J.R.; Tyrrell, D.A. Experimental parvoviral infection in humans. J. Infect. Dis. 1985, 152, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Ramsauer, A.S.; Badenhorst, M.; Cavalleri, J.-M.V. Equine parvovirus hepatitis. Equine Vet. J. 2021, 53, 886–894. [Google Scholar] [CrossRef]
- Ingrand, D. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Virologie. 2006, 10, 67–68. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Tse, H.; Fu, C.T.Y.; Au, W.-K.; Chen, X.-C.; Tsoi, H.-W.; Tsang, T.H.F.; Chan, J.S.Y.; Tsang, D.N.C.; et al. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 2008, 89, 1840–1848. [Google Scholar] [CrossRef]
- Xiao, C.-T.; Giménez-Lirola, L.G.; Jiang, Y.-H.; Halbur, P.G.; Opriessnig, T. Characterization of a novel porcine parvovirus tentatively designated PPV5. PLoS ONE 2013, 8, e65312. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Qiao, C.; Han, X.; Han, T.; Kang, W.; Zi, Z.; Cao, Z.; Zhai, X.; Cai, X. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol. J. 2014, 11, 203. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Xiao, C.-T.; Gerber, P.F.; Halbur, P.G. Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet. Microbiol. 2014, 173, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.M.; Mitra, N.; Hause, B.M. Discovery of a novel Parvovirinae virus, porcine parvovirus 7, by metagenomic sequencing of porcine rectal swabs. Virus Genes 2016, 52, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yan, G.; Chen, S.; Han, H.; Li, J.; Zhang, H.; Luo, S.; Liu, M.; Wu, Q.; Li, Q.; et al. Identification and genomic characterization of a novel porcine parvovirus in China. Front. Vet. Sci. 2022, 9, 1009103. [Google Scholar] [CrossRef] [PubMed]
- Streck, A.F.; Truyen, U. Porcine Parvovirus. Curr. Issues Mol. Biol. 2020, 37, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Novosel, D.; Cadar, D.; Tuboly, T.; Jungic, A.; Stadejek, T.; Ait-Ali, T.; Cságola, A. Investigating porcine parvoviruses genogroup 2 infection using in situ polymerase chain reaction. BMC Vet. Res. 2018, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Nelsen, A.; Lin, C.-M.; Hause, B.M. Porcine parvovirus 2 is predominantly associated with macrophages in porcine respiratory disease complex. Front. Vet. Sci. 2021, 8, 726884. [Google Scholar] [CrossRef]
- Ndze, V.N.; Cadar, D.; Cságola, A.; Kisfali, P.; Kovács, E.; Farkas, S.; Ngu, A.F.; Esona, M.D.; Dán, Á.; Tuboly, T.; et al. Detection of novel porcine bocaviruses in fecal samples of asymptomatic pigs in Cameroon. Infect. Genet. Evol. 2013, 17, 277–282. [Google Scholar] [CrossRef]
- Chaabouni, L.; Amira, C.; Abdelmoula, L.; Ben Had, Y.C.; Kchir, M.M.; Zouari, R. Rheumatologic manifestations of parvovirus B19 infection. Tunis. Med. 2004, 82, 642–647. [Google Scholar]
- Mietzsch, M.; Pénzes, J.J.; Agbandje-McKenna, M. Twenty-Five Years of Structural Parvovirology. Viruses 2019, 11, 362. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiao, Y.; Qiu, M.; Li, X.; Li, S.; Lin, H.; Li, X.; Zhu, J.; Chen, N. A Systematic Investigation Unveils High Coinfection Status of Porcine Parvovirus Types 1 through 7 in China from 2016 to 2020. Microbiol. Spectr. 2021, 9, e0129421. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Stadejek, T. The detection and genetic diversity of novel porcine parvovirus 7 (PPV7) on Polish pig farms. Res. Vet. Sci. 2018, 120, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Lagan Tregaskis, P.; Staines, A.; Gordon, A.; Sheridan, P.; McMenamy, M.; Duffy, C.; Collins, P.J.; Mooney, M.H.; Lemon, K. Co-infection status of novel parvovirus’s (PPV2 to 4) with porcine circovirus 2 in porcine respiratory disease complex and porcine circovirus-associated disease from 1997 to 2012. Transbound. Emerg. Dis. 2021, 68, 1979–1994. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Cságola, A.; Kiss, T.; Tuboly, T. Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Mol. Phylogenet. Evol. 2013, 66, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Tattersall, P. Parvoviruses: Small does not mean simple. Annu. Rev. Virol. 2014, 1, 517–537. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; de Souza, W.M.; Agbandje-McKenna, M.; Gifford, R.J. An Ancient Lineage of Highly Divergent Parvoviruses Infects both Vertebrate and Invertebrate Hosts. Viruses 2019, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sol, N.; Le Junter, J.; Vassias, I.; Freyssinier, J.M.; Thomas, A.; Prigent, A.F.; Rudkin, B.B.; Fichelson, S.; Morinet, F. Possible interactions between the NS-1 protein and tumor necrosis factor alpha pathways in erythroid cell apoptosis induced by human parvovirus B19. J. Virol. 1999, 73, 8762–8770. [Google Scholar] [CrossRef]
- Miller, C.L.; Pintel, D.J. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 2002, 76, 3257–3266. [Google Scholar] [CrossRef]
- Lin, W.; Qiu, Z.; Liu, Q.; Cui, S. Interferon induction and suppression in swine testicle cells by porcine parvovirus and its proteins. Vet. Microbiol. 2013, 163, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Zádori, Z.; Szelei, J.; Lacoste, M.C.; Li, Y.; Gariépy, S.; Raymond, P.; Allaire, M.; Nabi, I.R.; Tijssen, P. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 2001, 1, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.; Wu, G.; Wang, D.; Bayles, D.O.; Lager, K.M.; Vincent, A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010, 155, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Zádori, Z.; Szelei, J.; Tijssen, P. SAT: A late NS protein of porcine parvovirus. J. Virol. 2005, 79, 13129–13138. [Google Scholar] [CrossRef]
- Manteufel, J.; Truyen, U. Animal bocaviruses: A brief review. Intervirology 2008, 51, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, J.; Menezes, J.; Tijssen, P. Genomic organization and mapping of transcription and translation products of the NADL-2 strain of porcine parvovirus. Virology 1993, 197, 86–98. [Google Scholar] [CrossRef]
- Campos, F.S.; Kluge, M.; Franco, A.C.; Giongo, A.; Valdez, F.P.; Saddi, T.M.; Brito, W.M.E.D.; Roehe, P.M. Complete Genome Sequence of Porcine Parvovirus 2 Recovered from Swine Sera. Genome Announc. 2016, 4, e01627-15. [Google Scholar] [CrossRef]
- Xiao, C.-T.; Halbur, P.G.; Opriessnig, T. Complete genome sequence of a novel porcine parvovirus (PPV) provisionally designated PPV5. Genome Announc. 2013, 1, e00021-12. [Google Scholar] [CrossRef]
- Chung, H.-C.; Nguyen, V.-G.; Huynh, T.-M.-L.; Park, Y.-H.; Park, K.-T.; Park, B.-K. PCR-based detection and genetic characterization of porcine parvoviruses in South Korea in 2018. BMC Vet. Res. 2020, 16, 113. [Google Scholar] [CrossRef]
- Lou, S.; Xu, B.; Huang, Q.; Zhi, N.; Cheng, F.; Wong, S.; Brown, K.; Delwart, E.; Liu, Z.; Qiu, J. Molecular characterization of the newly identified human parvovirus 4 in the family Parvoviridae. Virology 2012, 422, 59–69. [Google Scholar] [CrossRef][Green Version]
- Bergeron, J.; Hébert, B.; Tijssen, P. Genome organization of the Kresse strain of porcine parvovirus: Identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J. Virol. 1996, 70, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.A.; Hébert, B.; Sullivan, G.M.; Parrish, C.R.; Zádori, Z.; Tijssen, P.; Rossmann, M.G. The structure of porcine parvovirus: Comparison with related viruses. J. Mol. Biol. 2002, 315, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.S.; Rossmann, M.G. Structure, sequence, and function correlations among parvoviruses. Virology 1993, 194, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, P.; Ward, D.C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 1976, 263, 106–109. [Google Scholar] [CrossRef] [PubMed]
- López-Bueno, A.; Villarreal, L.P.; Almendral, J.M. Parvovirus variation for disease: A difference with RNA viruses? Curr. Top. Microbiol. Immunol. 2006, 299, 349–370. [Google Scholar] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Cadar, D.; Dán, Á.; Tombácz, K.; Lőrincz, M.; Kiss, T.; Becskei, Z.; Spînu, M.; Tuboly, T.; Cságola, A. Phylogeny and evolutionary genetics of porcine parvovirus in wild boars. Infect. Genet. Evol. 2012, 12, 1163–1171. [Google Scholar] [CrossRef]
- Streck, A.F.; Bonatto, S.L.; Homeier, T.; Souza, C.K.; Gonçalves, K.R.; Gava, D.; Canal, C.W.; Truyen, U. High rate of viral evolution in the capsid protein of porcine parvovirus. J. Gen. Virol. 2011, 92, 2628–2636. [Google Scholar] [CrossRef]
- Zimmermann, P.; Ritzmann, M.; Selbitz, H.J.; Heinritzi, K.; Truyen, U. VP1 sequences of German porcine parvovirus isolates define two genetic lineages. J. Gen. Virol. 2006, 87, 295–301. [Google Scholar] [CrossRef]
- Oh, W.-T.; Kim, R.-Y.; Nguyen, V.-G.; Chung, H.-C.; Park, B.-K. Perspectives on the evolution of porcine parvovirus. Viruses 2017, 9, 196. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, N.; Kvisgaard, L.K.; Baele, G.; Boone, C.; Kunze, M.; Larsen, L.E.; Theuns, S.; Nauwynck, H. Molecular epidemiology of Porcine Parvovirus Type 1 (PPV1) and the reactivity of vaccine-induced antisera against historical and current PPV1 strains. Virus Evol. 2022, 8, veac053. [Google Scholar] [CrossRef] [PubMed]
- Streck, A.F.; Canal, C.W.; Truyen, U. Molecular epidemiology and evolution of porcine parvoviruses. Infect. Genet. Evol. 2015, 36, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Zeeuw, E.J.L.; Leinecker, N.; Herwig, V.; Selbitz, H.J.; Truyen, U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J. Gen. Virol. 2007, 88, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Lőrincz, M.; Kiss, T.; Novosel, D.; Podgorska, K.; Becskei, Z.; Tuboly, T.; Cságola, A. Emerging novel porcine parvoviruses in Europe: Origin, evolution, phylodynamics and phylogeography. J. Gen. Virol. 2013, 94, 2330–2337. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Mai, J.; Yang, Y.; Wang, N. Porcine Parvovirus 7: Evolutionary Dynamics and Identification of Epitopes toward Vaccine Design. Vaccines 2020, 8, 359. [Google Scholar] [CrossRef] [PubMed]
- Saekhow, P.; Mawatari, T.; Ikeda, H. Coexistence of multiple strains of porcine parvovirus 2 in pig farms. Microbiol. Immunol. 2014, 58, 382–387. [Google Scholar] [CrossRef]
- Sun, J.; Huang, L.; Wei, Y.; Wang, Y.; Chen, D.; Du, W.; Wu, H.; Liu, C. Prevalence of emerging porcine parvoviruses and their co-infections with porcine circovirus type 2 in China. Arch. Virol. 2015, 160, 1339–1344. [Google Scholar] [CrossRef]
- Amoroso, M.G.; Cerutti, F.; D’Alessio, N.; Lucibelli, M.G.; Cerrone, A.; Acutis, P.L.; Galiero, G.; Fusco, G.; Peletto, S. First identification of porcine parvovirus 3 in a wild boar in Italy by viral metagenomics—Short communication. Acta Vet. Hung. 2019, 67, 135–139. [Google Scholar] [CrossRef]
- Kim, S.-C.; Kim, J.-H.; Kim, J.-Y.; Park, G.-S.; Jeong, C.-G.; Kim, W.-I. Prevalence of porcine parvovirus 1 through 7 (PPV1-PPV7) and co-factor association with PCV2 and PRRSV in Korea. BMC Vet. Res. 2022, 18, 133. [Google Scholar] [CrossRef]
- Cui, J.; Fan, J.; Gerber, P.F.; Biernacka, K.; Stadejek, T.; Xiao, C.-T.; Opriessnig, T. First identification of porcine parvovirus 6 in Poland. Virus Genes 2017, 53, 100–104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vargas-Bermudez, D.S.; Rendon-Marin, S.; Ruiz-Saenz, J.; Mogollón, D.; Jaime, J. The first report of porcine parvovirus 7 (PPV7) in Colombia demonstrates the presence of variants associated with modifications at the level of the VP2-capsid protein. PLoS ONE 2021, 16, e0258311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zheng, C.; Lv, Z.; Xue, S.; Chen, Y.; Liu, Y.; Huang, X.; Luo, G.; Yang, X.; Dai, A. Genetic and epidemic characteristics of porcine parvovirus 7 in the Fujian and Guangdong regions of southern China. Front. Vet. Sci. 2022, 9, 949764. [Google Scholar] [CrossRef] [PubMed]
- Shackelton, L.A.; Hoelzer, K.; Parrish, C.R.; Holmes, E.C. Comparative analysis reveals frequent recombination in the parvoviruses. J. Gen. Virol. 2007, 88, 3294–3301. [Google Scholar] [CrossRef] [PubMed]
- Parsyan, A.; Szmaragd, C.; Allain, J.-P.; Candotti, D. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J. Gen. Virol. 2007, 88, 428–431. [Google Scholar] [CrossRef] [PubMed]
- Park, G.-N.; Song, S.; Cha, R.M.; Choe, S.; Shin, J.; Kim, S.-Y.; Hyun, B.-H.; Park, B.-K.; An, D.-J. Genetic analysis of porcine parvoviruses detected in South Korean wild boars. Arch. Virol. 2021, 166, 2249–2254. [Google Scholar] [CrossRef] [PubMed]
- Leal, É.; Villanova, F.E.; Lin, W.; Hu, F.; Liu, Q.; Liu, Y.; Cui, S. Interclade recombination in porcine parvovirus strains. J. Gen. Virol. 2012, 93, 2692–2704. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Mahnel, H. Further studies on the cultivation of swine plague virus in cell cultures with a cytopathogenic effect. Zentralbl. Bakteriol. Orig. 1966, 199, 399–407. [Google Scholar]
- Mészáros, I.; Olasz, F.; Cságola, A.; Tijssen, P.; Zádori, Z. Biology of Porcine Parvovirus (Ungulate parvovirus 1). Viruses 2017, 9, 393. [Google Scholar] [CrossRef]
- Hijikata, M.; Abe, K.; Win, K.M.; Shimizu, Y.K.; Keicho, N.; Yoshikura, H. Identification of new parvovirus DNA sequence in swine sera from Myanmar. Jpn. J. Infect. Dis. 2001, 54, 244–245. [Google Scholar]
- Wang, F.; Wei, Y.; Zhu, C.; Huang, X.; Xu, Y.; Yu, L.; Yu, X. Novel parvovirus sublineage in the family of Parvoviridae. Virus Genes 2010, 41, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Kim, E.-J.; Cho, I.-S.; Lee, K.-K.; Shin, Y.-K. Complete Genome Sequences of Porcine Parvovirus 2 Isolated from Swine in the Republic of Korea. Genome Announc. 2017, 5, e01738-16. [Google Scholar] [CrossRef] [PubMed]
- Cságola, A.; Lőrincz, M.; Cadar, D.; Tombácz, K.; Biksi, I.; Tuboly, T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 2012, 157, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Streck, A.F.; Homeier, T.; Foerster, T.; Fischer, S.; Truyen, U. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Arch. Virol. 2013, 158, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Afolabi, K.O.; Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Prevalence of porcine parvoviruses in some South African swine herds with background of porcine circovirus type 2 infection. Acta Trop. 2019, 190, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, T.R.; Chamings, A.; Alexandersen, S. Exploring the Cause of Diarrhoea and Poor Growth in 8-11-Week-Old Pigs from an Australian Pig Herd Using Metagenomic Sequencing. Viruses 2021, 13, 1608. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.-T.; Gerber, P.F.; Giménez-Lirola, L.G.; Halbur, P.G.; Opriessnig, T. Characterization of porcine parvovirus type 2 (PPV2) which is highly prevalent in the USA. Vet. Microbiol. 2013, 161, 325–330. [Google Scholar] [CrossRef]
- Garcia-Camacho, L.A.; Vargas-Ruiz, A.; Marin-Flamand, E.; Ramírez-Álvarez, H.; Brown, C. A retrospective study of DNA prevalence of porcine parvoviruses in Mexico and its relationship with porcine circovirus associated disease. Microbiol. Immunol. 2020, 64, 366–376. [Google Scholar] [CrossRef]
- Adlhoch, C.; Kaiser, M.; Ellerbrok, H.; Pauli, G. High prevalence of porcine Hokovirus in German wild boar populations. Virol. J. 2010, 7, 171. [Google Scholar] [CrossRef]
- Cui, J.; Biernacka, K.; Fan, J.; Gerber, P.F.; Stadejek, T.; Opriessnig, T. Circulation of Porcine Parvovirus Types 1 through 6 in Serum Samples Obtained from Six Commercial Polish Pig Farms. Transbound. Emerg. Dis. 2017, 64, 1945–1952. [Google Scholar] [CrossRef]
- Sliz, I.; Vlasakova, M.; Jackova, A.; Vilcek, S. Characterization of porcine parvovirus type 3 and porcine circovirus type 2 in wild boars (Sus scrofa) in slovakia. J. Wildl. Dis. 2015, 51, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, Y.; Liu, J.; Tang, Q.; Liu, C. Prevalence of porcine hokovirus and its co-infection with porcine circovirus 2 in China. Arch. Virol. 2013, 158, 1987–1991. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.-T.; Giménez-Lirola, L.G.; Halbur, P.G.; Opriessnig, T. Increasing porcine PARV4 prevalence with pig age in the U.S. pig population. Vet. Microbiol. 2012, 160, 290–296. [Google Scholar] [CrossRef]
- Bellehumeur, C.; Boyle, B.; Mandeville, I.; Gagnon, C.A. High-throughput sequencing revealed the presence of an unforeseen parvovirus species in Canadian swine: The porcine partetravirus. Can. Vet. J. 2013, 54, 787–789. [Google Scholar] [PubMed]
- Souza, C.K.; Streck, A.F.; Gonçalves, K.R.; Pinto, L.D.; Ravazzolo, A.P.; de Barcellos, D.E.D.S.N.; Canal, C.W. Phylogenetic characterization of the first Ungulate tetraparvovirus 2 detected in pigs in Brazil. Braz. J. Microbiol 2016, 47, 513–517. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miranda, C.; Coelho, C.; Vieira-Pinto, M.; Thompson, G. Porcine hokovirus in wild boar in Portugal. Arch. Virol. 2016, 161, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Blomström, A.-L.; Belák, S.; Fossum, C.; McKillen, J.; Allan, G.; Wallgren, P.; Berg, M. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009, 146, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhai, S.-L.; Cheung, A.K.; Zhang, H.-B.; Long, J.-X.; Yuan, S.-S. Detection of a novel porcine parvovirus, PPV4, in Chinese swine herds. Virol. J. 2010, 7, 333. [Google Scholar] [CrossRef]
- Blomström, A.-L.; Ståhl, K.; Masembe, C.; Okoth, E.; Okurut, A.R.; Atmnedi, P.; Kemp, S.; Bishop, R.; Belák, S.; Berg, M. Viral metagenomic analysis of bushpigs (Potamochoerus larvatus) in Uganda identifies novel variants of Porcine parvovirus 4 and Torque teno sus virus 1 and 2. Virol. J. 2012, 9, 192. [Google Scholar] [CrossRef]
- Gava, D.; Souza, C.K.; Schaefer, R.; Vincent, A.L.; Cantão, M.E.; Coldebella, A.; Ciacci-Zanella, J.R. A TaqMan-based real-time PCR for detection and quantification of porcine parvovirus 4. J. Virol. Methods 2015, 219, 14–17. [Google Scholar] [CrossRef]
- Wu, R.; Wen, Y.; Huang, X.; Wen, X.; Yan, Q.; Huang, Y.; Ma, X.; Cao, S. First complete genomic characterization of a porcine parvovirus 5 isolate from China. Arch. Virol. 2014, 159, 1533–1536. [Google Scholar] [CrossRef] [PubMed]
- Cibulski, S.; Alves de Lima, D.; Fernandes Dos Santos, H.; Teixeira, T.F.; Tochetto, C.; Mayer, F.Q.; Roehe, P.M. A plate of viruses: Viral metagenomics of supermarket chicken, pork and beef from Brazil. Virology 2021, 552, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Schirtzinger, E.E.; Suddith, A.W.; Hause, B.M.; Hesse, R.A. First identification of porcine parvovirus 6 in North America by viral metagenomic sequencing of serum from pigs infected with porcine reproductive and respiratory syndrome virus. Virol. J. 2015, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Kekarainen, T.; Llorens, A.; Correa-Fiz, F.; Segalés, J. Exploratory metagenomic analyses of periweaning failure-to-thrive syndrome-affected pigs. Vet. Rec. 2019, 184, 25. [Google Scholar] [CrossRef] [PubMed]
- Miłek, D.; Woźniak, A.; Guzowska, M.; Stadejek, T. Detection Patterns of Porcine Parvovirus (PPV) and Novel Porcine Parvoviruses 2 through 6 (PPV2-PPV6) in Polish Swine Farms. Viruses 2019, 11, 474. [Google Scholar] [CrossRef]
- Xing, X.; Zhou, H.; Tong, L.; Chen, Y.; Sun, Y.; Wang, H.; Zhang, G. First identification of porcine parvovirus 7 in China. Arch. Virol. 2018, 163, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Ruan, W.; Yue, H.; Tang, C.; Zhou, K.; Zhang, B. Viral communities associated with porcine respiratory disease complex in intensive commercial farms in Sichuan province, China. Sci. Rep. 2018, 8, 13341. [Google Scholar] [CrossRef]
- Kim, S.-C.; Jeong, C.-G.; Nazki, S.; Lee, S.-I.; Baek, Y.-C.; Jung, Y.-J.; Kim, W.-I. Evaluation of a multiplex PCR method for the detection of porcine parvovirus types 1 through 7 using various field samples. PLoS ONE 2021, 16, e0245699. [Google Scholar] [CrossRef]
- Da Silva, M.S.; Budaszewski, R.F.; Weber, M.N.; Cibulski, S.P.; Paim, W.P.; Mósena, A.C.S.; Canova, R.; Varela, A.P.M.; Mayer, F.Q.; Pereira, C.W.; et al. Liver virome of healthy pigs reveals diverse small ssDNA viral genomes. Infect. Genet. Evol. 2020, 81, 104203. [Google Scholar] [CrossRef]
- Ouh, I.-O.; Park, S.; Lee, J.-Y.; Song, J.Y.; Cho, I.-S.; Kim, H.-R.; Park, C.-K. First detection and genetic characterization of porcine parvovirus 7 from Korean domestic pig farms. J. Vet. Sci. 2018, 19, 855–857. [Google Scholar] [CrossRef]
- Choi, C.S.; Molitor, T.W.; Joo, H.S.; Gunther, R. Pathogenicity of a skin isolate of porcine parvovirus in swine fetuses. Vet. Microbiol. 1987, 15, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, L.; Deng, X.; Kapusinszky, B.; Delwart, E. What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology 2014, 468–470, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Cutler, R.; Molitor, T.W.; Sauber, T.E.; Leman, A.D. Role of the rat in the transmission of porcine parvovirus. Am. J. Vet. Res. 1982, 43, 493–496. [Google Scholar] [PubMed]
- Linden, A.; Gilliaux, G.; Paternostre, J.; Benzarti, E.; Rivas, J.F.; Desmecht, D.; Garigliany, M. A novel parvovirus, Roe deer copiparvovirus, identified in Ixodes ricinus ticks. Virus Genes 2019, 55, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, D.S.; Howard, K.I.; Reuterman, N.A. Multivariate analysis of gang delinquency: III. age and physique of gangs and clubs. Multivar. Behav. Res. 1971, 6, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.S.; Donaldson-Wood, C.R.; Johnson, R.H. A standardised haemagglutination inhibition test for porcine parvovirus antibody. Aust. Vet. J. 1976, 52, 422–424. [Google Scholar] [CrossRef] [PubMed]
- Jeoung, H.-Y.; Lim, S.-I.; Kim, J.-J.; Cho, Y.-Y.; Kim, Y.K.; Song, J.-Y.; Hyun, B.-H.; An, D.-J. Serological prevalence of viral agents that induce reproductive failure in South Korean wild boar. BMC Vet. Res. 2015, 11, 78. [Google Scholar] [CrossRef]
- Song, C.; Zhu, C.; Zhang, C.; Cui, S. Detection of porcine parvovirus using a taqman-based real-time pcr with primers and probe designed for the NS1 gene. Virol. J. 2010, 7, 353. [Google Scholar] [CrossRef]
- Streck, A.F.; Hergemöller, F.; Rüster, D.; Speck, S.; Truyen, U. A TaqMan qPCR for quantitation of Ungulate protoparvovirus 1 validated in several matrices. J. Virol. Methods 2015, 218, 46–50. [Google Scholar] [CrossRef]
- Yu, H.-Q.; Cai, X.-Q.; Lin, Z.-X.; Li, X.-L.; Yue, Q.-Y.; Li, R.; Zhu, X.-Q. Rapid and specific detection of porcine parvovirus using real-time PCR and high resolution melting (HRM) analysis. BMC Vet. Res. 2015, 11, 46. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Yang, S.; Ma, L.; Ma, Y.; Liu, X.; Cai, X.; Zhang, Y.; Liu, Y. Rapid detection of porcine parvovirus DNA by sensitive loop-mediated isothermal amplification. J. Virol. Methods 2009, 158, 100–103. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, W.; Fan, J.; Fan, S.; Ding, H.; Chen, J.; Yi, L. Recombinase-Aided Amplification Coupled with Lateral Flow Dipstick for Efficient and Accurate Detection of Porcine Parvovirus. Life 2021, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, S.F.; Lucas, M.; Huck, R.A. A small haemagglutinating porcine DNA virus. I. Isolation and properties. J. Comp. Pathol. 1969, 79, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, M.; Bouchard-Lévesque, V.; Fernandes, S.; Tijssen, P. Classic nuclear localization signals and a novel nuclear localization motif are required for nuclear transport of porcine parvovirus capsid proteins. J. Virol. 2014, 88, 11748–11759. [Google Scholar] [CrossRef]
- Heldt, C.L.; Hernandez, R.; Mudiganti, U.; Gurgel, P.V.; Brown, D.T.; Carbonell, R.G. A colorimetric assay for viral agents that produce cytopathic effects. J. Virol. Methods 2006, 135, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.S.; Donaldson-Wood, C.R.; Johnson, R.H. Observations on the pathogenesis of porcine parvovirus infection. Arch. Virol. 1976, 51, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef]
- Cságola, A.; Zádori, Z.; Mészáros, I.; Tuboly, T. Detection of porcine parvovirus 2 (ungulate tetraparvovirus 3) specific antibodies and examination of the serological profile of an infected swine herd. PLoS ONE 2016, 11, e0151036. [Google Scholar] [CrossRef]
- Miłek, D.; Woźniak, A.; Podgórska, K.; Stadejek, T. Do porcine parvoviruses 1 through 7 (PPV1-PPV7) have an impact on porcine circovirus type 2 (PCV2) viremia in pigs? Vet. Microbiol. 2020, 242, 108613. [Google Scholar] [CrossRef]
- Thuy, N.T.D.; Trung, N.T.; Dung, T.Q.; Khoa, D.V.A.; Thuy, D.T.N.; Opriessnig, T. First investigation of the prevalence of parvoviruses in slaughterhouse pigs and genomic characterization of ungulate copiparvovirus 2 in Vietnam. Arch. Virol. 2021, 166, 779–788. [Google Scholar] [CrossRef]
- Saekhow, P.; Ikeda, H. Prevalence and genomic characterization of porcine parvoviruses detected in Chiangmai area of Thailand in 2011. Microbiol. Immunol. 2015, 59, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Xiao, C.-T.; Gerber, P.F.; Halbur, P.G. Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the U.S. concurrently infected with PPV2. Vet. Microbiol. 2013, 163, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Bisimwa, P.N.; Wasso, D.S.; Bantuzeko, F.; Aksanti, C.B.; Tonui, R.; Birindwa, A.B.; Bisimwa, E.B. First investigation on the presence of porcine parvovirus type 3 in domestic pig farms without reproductive failure in the Democratic Republic of Congo. Vet. Anim. Sci. 2021, 13, 100187. [Google Scholar] [CrossRef] [PubMed]
- Cibulski, S.P.; Teixeira, T.F.; Varela, A.P.M.; Scheffer, C.M.; Santos, H.F.; Lima, F.E.S.; Roehe, P.M. Ungulate copiparvovirus 2 in healthy and postweaning multisystemic wasting syndrome-affected pigs. Trop. Anim. Health Prod. 2017, 49, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Komina, A.; Anoyatbekova, A.; Krasnikov, N.; Yuzhakov, A. Identification and in vitro characterization of a novel porcine parvovirus 6 in Russia. Vet. Res. Commun. 2023, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, K.-K.; Wang, J.; Wang, X.-P.; Zhao, L.; Sun, P.; Li, Y.-D. Detection and molecular characterization of novel porcine parvovirus 7 in Anhui province from Central-Eastern China. Infect. Genet. Evol. 2019, 71, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Pescador, C.A.; Bandarra, P.M.; Castro, L.A.; Antoniassi, N.A.B.; Ravazzolo, A.P.; Sonne, L.; Cruz, C.E.F.; Driemeier, D. Co-infection by porcine circovirus type 2 and porcine parvovirus in aborted fetuses and stillborn piglets in southern Brazil. Pesq. Vet. Bras. 2007, 27, 425–429. [Google Scholar] [CrossRef]
- Kresse, J.I.; Taylor, W.D.; Stewart, W.W.; Eernisse, K.A. Parvovirus infection in pigs with necrotic and vesicle-like lesions. Vet. Microbiol. 1985, 10, 525–531. [Google Scholar] [CrossRef]
- Saade, G.; Deblanc, C.; Bougon, J.; Marois-Créhan, C.; Fablet, C.; Auray, G.; Belloc, C.; Leblanc-Maridor, M.; Gagnon, C.A.; Zhu, J.; et al. Coinfections and their molecular consequences in the porcine respiratory tract. Vet. Res. 2020, 51, 80. [Google Scholar] [CrossRef]
- Opriessnig, T.; Fenaux, M.; Yu, S.; Evans, R.B.; Cavanaugh, D.; Gallup, J.M.; Pallares, F.J.; Thacker, E.L.; Lager, K.M.; Meng, X.J.; et al. Effect of porcine parvovirus vaccination on the development of PMWS in segregated early weaned pigs coinfected with type 2 porcine circovirus and porcine parvovirus. Vet. Microbiol. 2004, 98, 209–220. [Google Scholar] [CrossRef]
- Allan, G.M.; Kennedy, S.; McNeilly, F.; Foster, J.C.; Ellis, J.A.; Krakowka, S.J.; Meehan, B.M.; Adair, B.M. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 1999, 121, 1–11. [Google Scholar] [CrossRef]
- Kennedy, S.; Moffett, D.; McNeilly, F.; Meehan, B.; Ellis, J.; Krakowka, S.; Allan, G.M. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 2000, 122, 9–24. [Google Scholar] [CrossRef]
- Sharma, R.; Saikumar, G. Porcine parvovirus- and porcine circovirus 2-associated reproductive failure and neonatal mortality in crossbred Indian pigs. Trop. Anim. Health Prod. 2010, 42, 515–522. [Google Scholar] [CrossRef]
- Opriessnig, T.; Karuppannan, A.K.; Halbur, P.G.; Calvert, J.G.; Nitzel, G.P.; Matzinger, S.R.; Meng, X.-J. Porcine circovirus type 2a or 2b based experimental vaccines provide protection against PCV2d/porcine parvovirus 2 co-challenge. Vaccine 2020, 38, 1975–1981. [Google Scholar] [CrossRef]
- Wang, W.; Cao, L.; Sun, W.; Xin, J.; Zheng, M.; Tian, M.; Lu, H.; Jin, N. Sequence and phylogenetic analysis of novel porcine parvovirus 7 isolates from pigs in Guangxi, China. PLoS ONE 2019, 14, e0219560. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Bermudez, D.S.; Mogollon, J.D.; Franco-Rodriguez, C.; Jaime, J. The Novel Porcine Parvoviruses: Current State of Knowledge and Their Possible Implications in Clinical Syndromes in Pigs. Viruses 2023, 15, 2398. https://doi.org/10.3390/v15122398
Vargas-Bermudez DS, Mogollon JD, Franco-Rodriguez C, Jaime J. The Novel Porcine Parvoviruses: Current State of Knowledge and Their Possible Implications in Clinical Syndromes in Pigs. Viruses. 2023; 15(12):2398. https://doi.org/10.3390/v15122398
Chicago/Turabian StyleVargas-Bermudez, Diana S., Jose Dario Mogollon, Camila Franco-Rodriguez, and Jairo Jaime. 2023. "The Novel Porcine Parvoviruses: Current State of Knowledge and Their Possible Implications in Clinical Syndromes in Pigs" Viruses 15, no. 12: 2398. https://doi.org/10.3390/v15122398
APA StyleVargas-Bermudez, D. S., Mogollon, J. D., Franco-Rodriguez, C., & Jaime, J. (2023). The Novel Porcine Parvoviruses: Current State of Knowledge and Their Possible Implications in Clinical Syndromes in Pigs. Viruses, 15(12), 2398. https://doi.org/10.3390/v15122398








