Nucleolin Regulates the Expression of Kaposi’s Sarcoma-Associated Herpesvirus’ Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA
Abstract
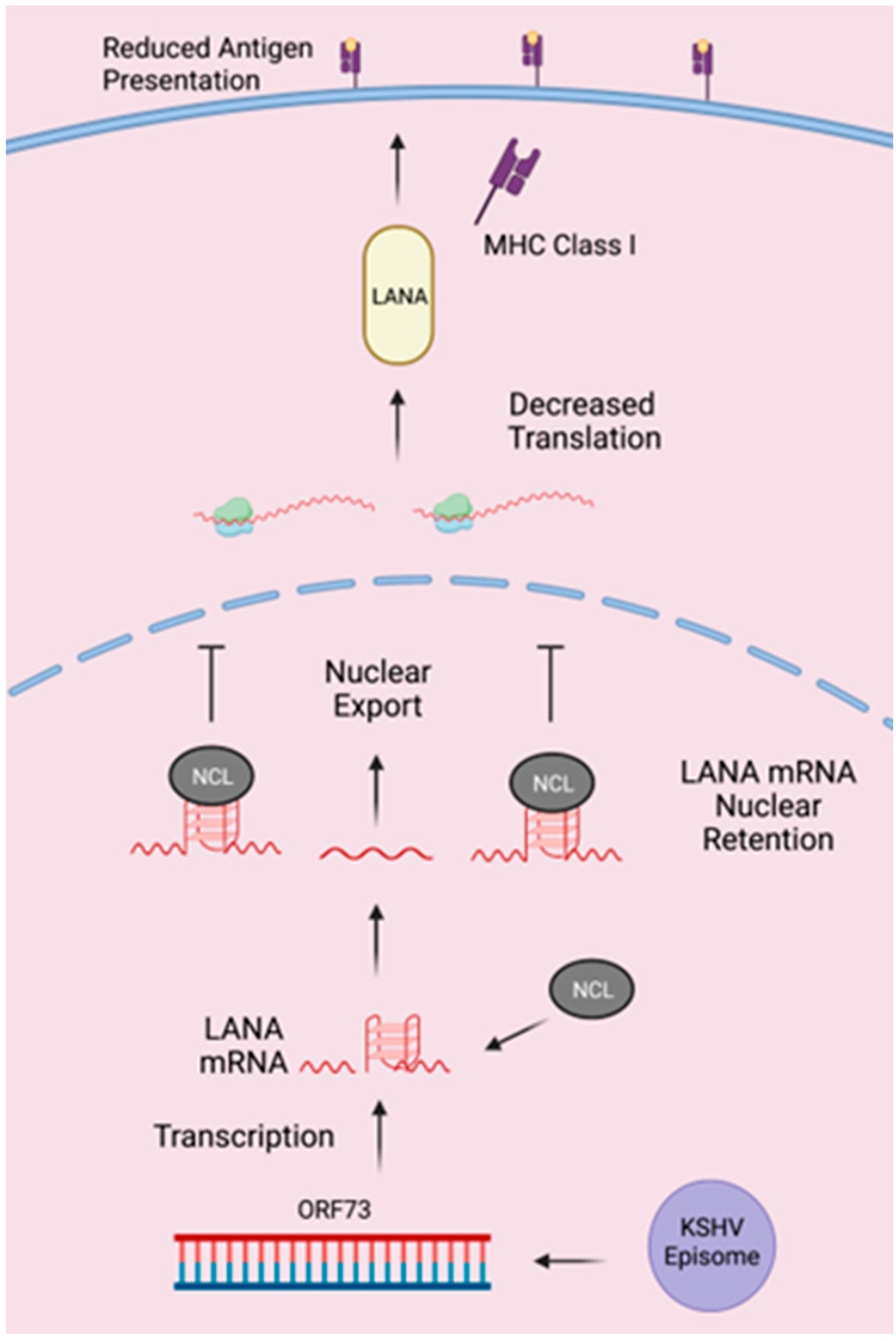
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Dual Luciferase Assay
2.3. Fluorescent In Situ Hybridization (FISH)
2.4. Antigen Presentation Assay
2.5. RNA Cross-Linking Immunoprecipitation Assay (RNA-CLIP)
2.6. In Vitro RNA Pulldown Assay
2.7. Fractionation and qRT-PCR
2.8. Statistical Analysis
2.9. Figure Generation
3. Results
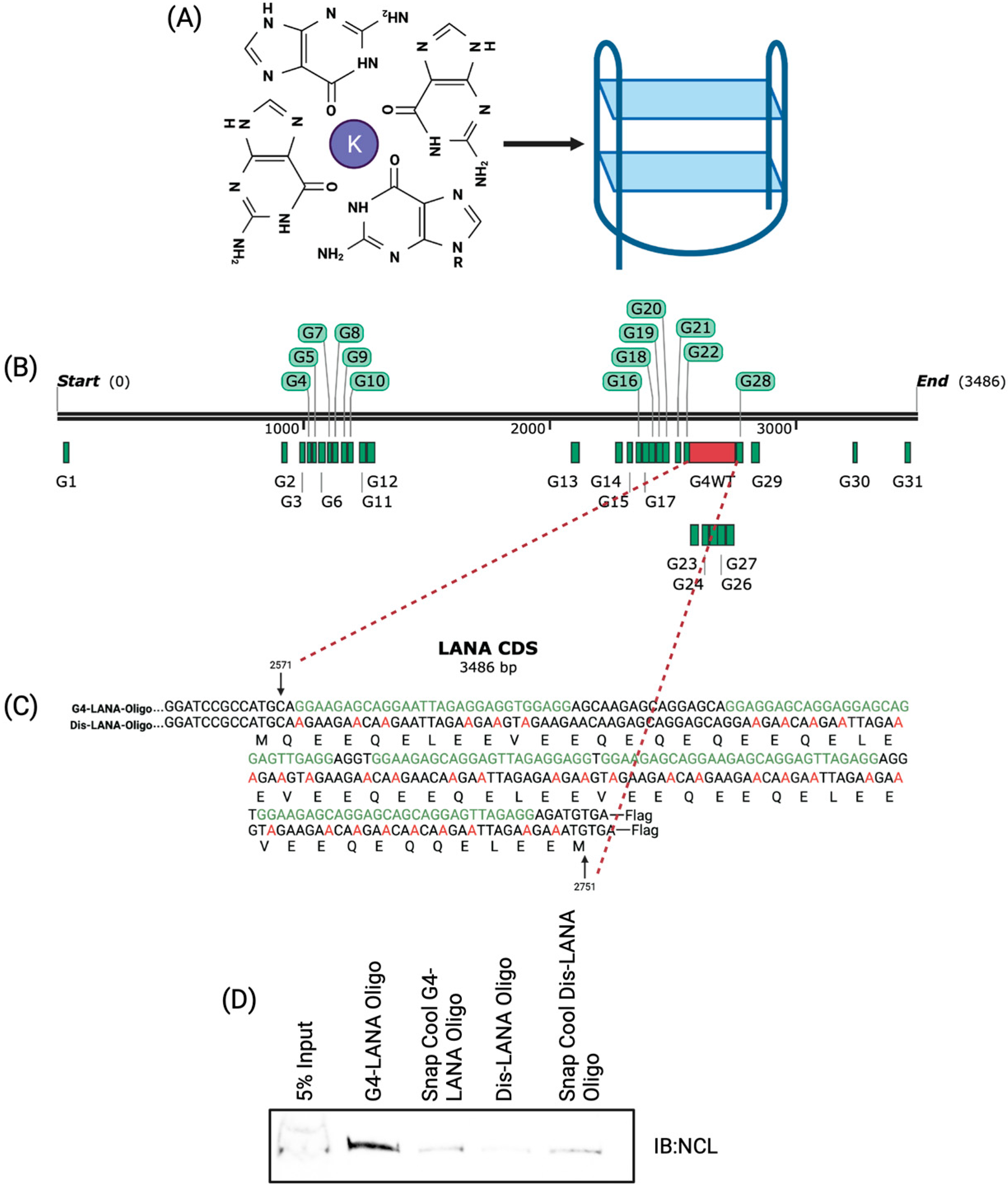
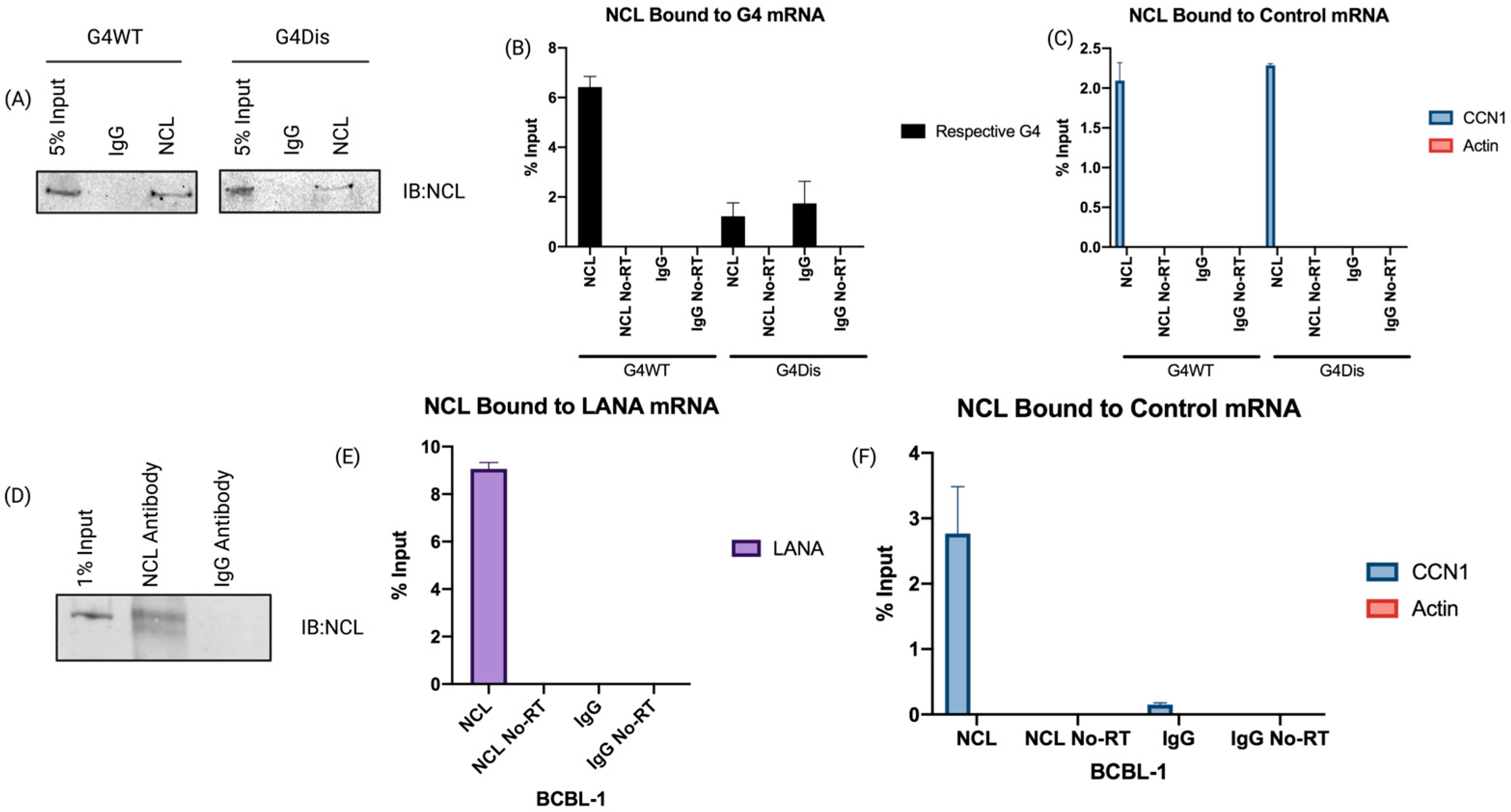
3.1. Nucleolin Binds to the G-Quadruplex of LANA mRNA
3.2. Increasing Expression of NCL Inhibited LANA Translation
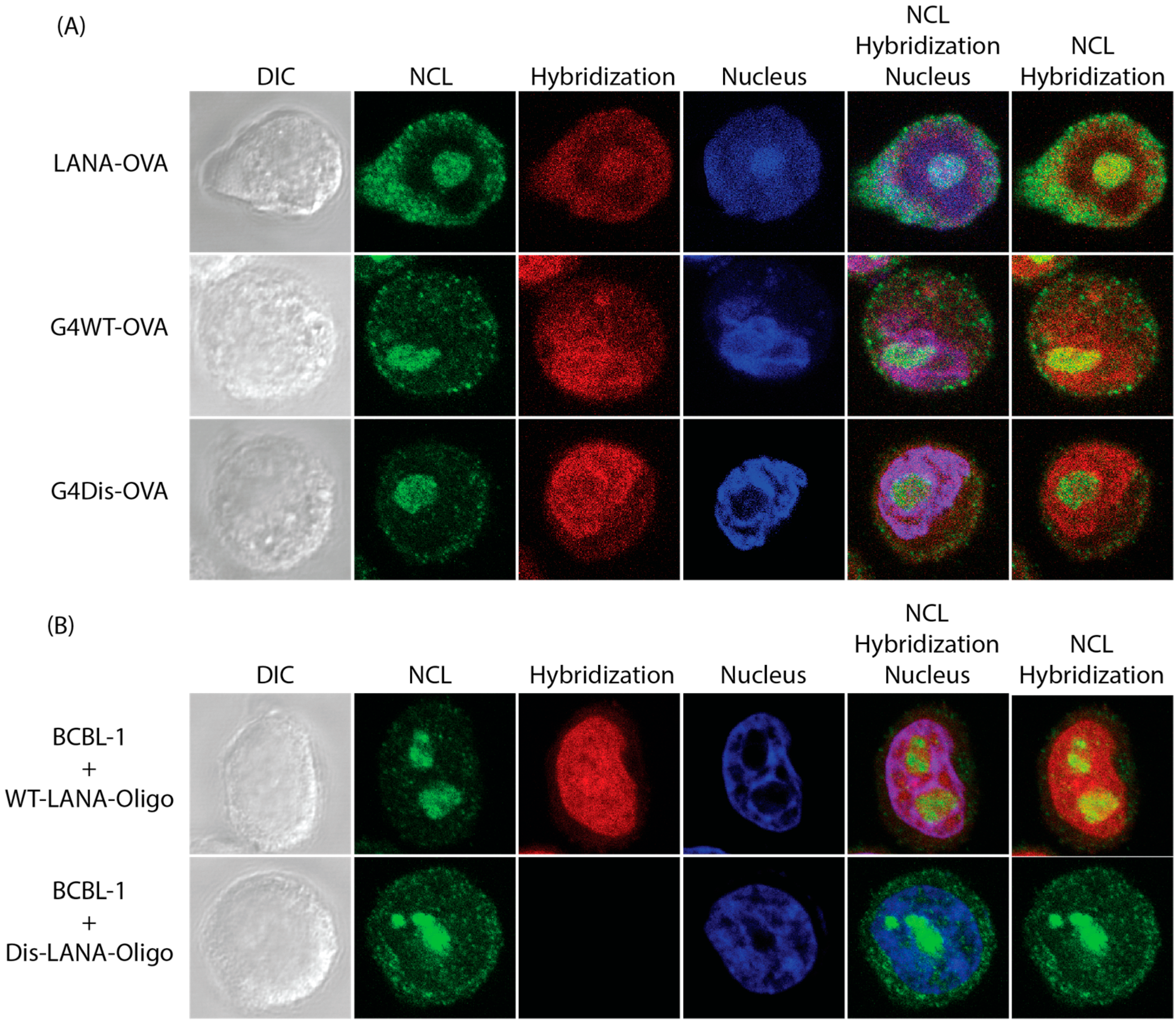
3.3. LANA mRNA Colocalizes with NCL
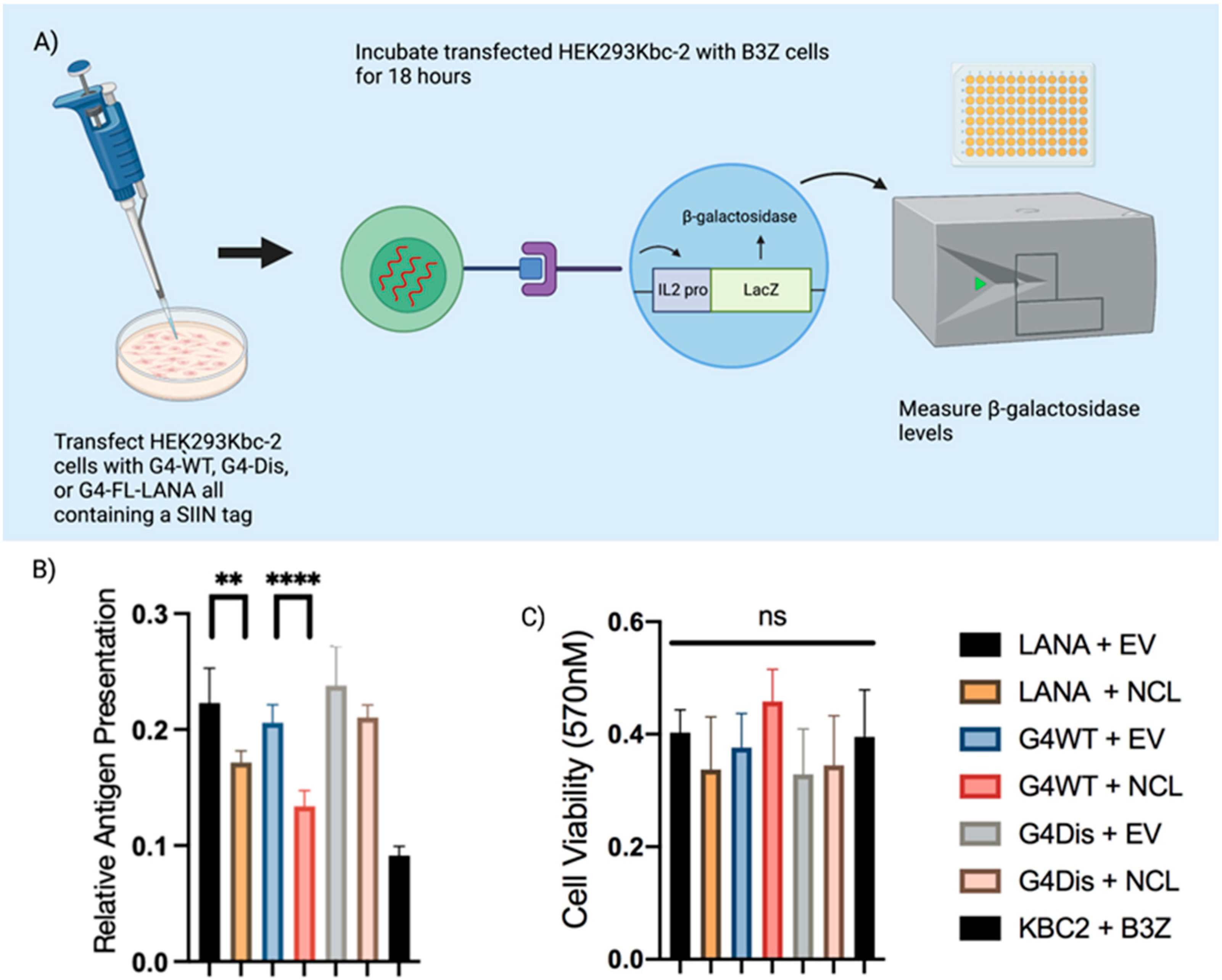
3.4. Interaction of LANA mRNA with NCL Decreases Antigen Presentation
3.5. Downregulation of Nucleolin Resulted in an Altered LANA mRNA Localization and Increased LANA Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin Melissa, S.; Lee, F.; Culpepper, J.; Knowles Daniel, M.; Moore Patrick, S. Identification of Herpesvirus-Like DNA Sequences in AIDS-Sssociated Kaposi’s Sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, E.; Chang, Y.; Moore, P.S.; Said, J.W.; Knowles, D.M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 1995, 332, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d’Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L.; et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995, 86, 1276–1280. [Google Scholar] [CrossRef]
- Nador, R.G.; Cesarman, E.; Chadburn, A.; Dawson, D.B.; Ansari, M.Q.; Sald, J.; Knowles, D.M. Primary effusion lymphoma: A distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 1996, 88, 645–656. [Google Scholar] [CrossRef]
- Renne, R.; Zhong, W.; Herndier, B.; McGrath, M.; Abbey, N.; Kedes, D.; Ganem, D. Lytic growth of Kaposi’s sarcoma–associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996, 2, 342–346. [Google Scholar] [CrossRef]
- Decker, L.L.; Shankar, P.; Khan, G.; Freeman, R.B.; Dezube, B.J.; Lieberman, J.; Thorley-Lawson, D.A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 1996, 184, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Ballestas, M.E.; Kaye, K.M. The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Future Microbiol. 2011, 6, 1399–1413. [Google Scholar] [CrossRef]
- Kedes, D.H.; Lagunoff, M.; Renne, R.; Ganem, D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J. Clin. Investig. 1997, 100, 2606–2610. [Google Scholar] [CrossRef]
- Rezaee, S.A.R.; Cunningham, C.; Davison, A.J.; Blackbourn, D.J. Kaposi’s sarcoma-associated herpesvirus immune modulation: An overview. J. Gen. Virol. 2006, 87, 1781–1804. [Google Scholar] [CrossRef]
- Kwun, H.J.; da Silva, S.R.; Qin, H.; Ferris, R.L.; Tan, R.; Chang, Y.; Moore, P.S. The central repeat domain 1 of Kaposi’s sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology 2011, 412, 357–365. [Google Scholar] [CrossRef]
- Zaldumbide, A.; Ossevoort, M.; Wiertz, E.J.H.J.; Hoeben, R.C. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma Herpes virus. Mol. Immunol. 2007, 44, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Sorel, O.; Chen, T.; Myster, F.; Javaux, J.; Vanderplasschen, A.; Dewals, B.G. Macavirus latency-associated protein evades immune detection through regulation of protein synthesis in cis depending upon its glycin/glutamate-rich domain. PLoS Pathog. 2017, 13, e1006691. [Google Scholar] [CrossRef] [PubMed]
- Thakker, S.; Purushothaman, P.; Gupta, N.; Challa, S.; Cai, Q.; Verma Subhash, C. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Inhibits Major Histocompatibility Complex Class II Expression by Disrupting Enhanceosome Assembly through Binding with the Regulatory Factor X Complex. J. Virol. 2015, 89, 5536–5556. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Dhakal, S.; Cui, Y.; Koirala, D.; Ghimire, C.; Kushwaha, S.; Yu, Z.; Yangyuoru, P.M.; Mao, H. Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 2013, 41, 3915–3923. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Pandey, S.; Agarwala, P.; Maiti, S. Effect of Loops and G-Quartets on the Stability of RNA G-Quadruplexes. J. Phys. Chem. B 2013, 117, 6896–6905. [Google Scholar] [CrossRef]
- Millevoi, S.; Moine, H.; Vagner, S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA 2012, 3, 495–507. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Bugaut, A.; Kumari, S.; Balasubramanian, S. G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 2008, 36, 6260–6268. [Google Scholar] [CrossRef]
- Song, J.; Perreault, J.P.; Topisirovic, I.; Richard, S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation 2016, 4, e1244031. [Google Scholar] [CrossRef] [PubMed]
- Biswas, B.; Kandpal, M.; Jauhari, U.K.; Vivekanandan, P. Genome-wide analysis of G-quadruplexes in herpesvirus genomes. BMC Genom. 2016, 17, 949. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palu, G.; Richter, S.N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef] [PubMed]
- Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M.A.; Palu, G.; Flamand, L.; Calistri, A.; Richter, S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antivir. Res. 2015, 118, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Martins, R.P.; Billant, O.; Contesse, M.A.; Findakly, S.; Pochard, P.; Daskalogianni, C.; Beauvineau, C.; Guetta, C.; Jamin, C.; et al. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8, 16043. [Google Scholar] [CrossRef] [PubMed]
- Madireddy, A.; Purushothaman, P.; Loosbroock, C.P.; Robertson, E.S.; Schildkraut, C.L.; Verma, S.C. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 2016, 44, 3675–3694. [Google Scholar] [CrossRef]
- Dabral, P.; Babu, J.; Zareie, A.; Verma, S.C. LANA and hnRNP A1 Regulate the Translation of LANA mRNA through G-Quadruplexes. J. Virol. 2020, 94, e01508-19. [Google Scholar] [CrossRef]
- Blake, N. Immune evasion by gammaherpesvirus genome maintenance proteins. J. Gen. Virol. 2010, 91 Pt 4, 829–846. [Google Scholar] [CrossRef]
- Daskalogianni, C.; Pyndiah, S.; Apcher, S.; Mazars, A.; Manoury, B.; Ammari, N.; Nylander, K.; Voisset, C.; Blondel, M.; Fåhraeus, R. Epstein-Barr virus-encoded EBNA1 and ZEBRA: Targets for therapeutic strategies against EBV-carrying cancers. J. Pathol. 2015, 235, 334–341. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Tosoni, E.; Frasson, I.; Scalabrin, M.; Perrone, R.; Butovskaya, E.; Nadai, M.; Palu, G.; Fabris, D.; Richter, S.N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015, 43, 8884–8897. [Google Scholar] [CrossRef] [PubMed]
- González, V.; Guo, K.; Hurley, L.; Sun, D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635. [Google Scholar] [CrossRef] [PubMed]
- Puig Lombardi, E.; Londoño-Vallejo, A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020, 48, 1603. [Google Scholar] [CrossRef]
- Kharel, P.; Balaratnam, S.; Beals, N.; Basu, S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip. Rev. RNA 2020, 11, e1568. [Google Scholar] [CrossRef]
- Novoseltseva, A.A.; Ivanov, N.M.; Novikov, R.A.; Tkachev, Y.V.; Bunin, D.A.; Gambaryan, A.S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. Structural and Functional Aspects of G-Quadruplex Aptamers Which Bind a Broad Range of Influenza A Viruses. Biomolecules 2020, 10, 119. [Google Scholar] [CrossRef]
- Panera, N.; Tozzi, A.E.; Alisi, A. The G-Quadruplex/Helicase World as a Potential Antiviral Approach Against COVID-19. Drugs 2020, 80, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Nazia, P.; Shamim, A.; Cho, S.; Kim, K.K. Computational Approaches to Predict the Non-canonical DNAs. Curr. Bioinform. 2019, 14, 470–479. [Google Scholar] [CrossRef]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef]
- Maizels, N. G4-associated human diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef]
- Ou, T.M.; Lu, Y.J.; Tan, J.H.; Huang, Z.S.; Wong, K.Y.; Gu, L.Q. G-quadruplexes: Targets in anticancer drug design. ChemMedChem 2008, 3, 690–713. [Google Scholar] [CrossRef]
- White, E.W.; Tanious, F.; Ismail, M.A.; Reszka, A.P.; Neidle, S.; Boykin, D.W.; Wilson, W.D. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys. Chem. 2007, 126, 140–153. [Google Scholar] [CrossRef]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Cai, Q.; Kreider, E.; Lu, J.; Robertson, E.S. Comprehensive Analysis of LANA Interacting Proteins Essential for Viral Genome Tethering and Persistence. PLoS ONE 2013, 8, e74662. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; McDowell, M.E.; McGuinness, J.; Salas, R.; Rumjahn, S.M.; Verma, S.C. Kaposi’s sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats. J. Virol. 2012, 86, 9983–9994. [Google Scholar] [CrossRef]
- Kochan, J.; Wawro, M.; Kasza, A. Simultaneous detection of mRNA and protein in single cells using immunofluorescence-combined single-molecule RNA FISH. Biotechniques 2015, 59, 209–212, 214, 216 passim. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA Associates with Demethylases UTX and JMJD3 to Activate Lytic Replication through a Physical Interaction with the Virus Genome. PLOS Pathog. 2012, 8, e1002680. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; CordeliÈRes, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Bobbin, M.L.; Rossi, J.J. RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Annu. Rev. Pharm. Toxicol. 2016, 56, 103–122. [Google Scholar] [CrossRef]
- Lama, L.; Seidl, C.I.; Ryan, K. New insights into the promoterless transcription of DNA coligo templates by RNA polymerase III. Transcription 2014, 5, e27913. [Google Scholar] [CrossRef] [PubMed]
- Paddison, P.J.; Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef]
- Purushothaman, P.; Dabral, P.; Gupta, N.; Sarkar, R.; Verma, S.C. KSHV Genome Replication and Maintenance. Front. Microbiol. 2016, 7, 54. [Google Scholar] [CrossRef]
- De Leo, A.; Deng, Z.; Vladimirova, O.; Chen, H.-S.; Dheekollu, J.; Calderon, A.; Myers, K.A.; Hayden, J.; Keeney, F.; Kaufer, B.B.; et al. LANA oligomeric architecture is essential for KSHV nuclear body formation and viral genome maintenance during latency. PLOS Pathog. 2019, 15, e1007489. [Google Scholar] [CrossRef]
- Renne, R.; Barry, C.; Dittmer, D.; Compitello, N.; Brown Patrick, O.; Ganem, D. Modulation of Cellular and Viral Gene Expression by the Latency-Associated Nuclear Antigen of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2001, 75, 458–468. [Google Scholar] [CrossRef]
- Jeong, J.H.; Orvis, J.; Kim, J.W.; McMurtrey, C.P.; Renne, R.; Dittmer, D.P. Regulation and autoregulation of the promoter for the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J. Biol. Chem. 2004, 279, 16822–16831. [Google Scholar] [CrossRef]
- Kwun Hyun, J.; da Silva Suzane, R.; Shah Ishita, M.; Blake, N.; Moore Patrick, S.; Chang, Y. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen 1 Mimics Epstein-Barr Virus EBNA1 Immune Evasion through Central Repeat Domain Effects on Protein Processing. J. Virol. 2007, 81, 8225–8235. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S.; Read, M.A. G-quadruplexes as therapeutic targets. Biopolymers 2000, 56, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Tominaga, K.; Lee, E.K.; Srikantan, S.; Kang, M.J.; Kim, M.M.; Selimyan, R.; Martindale, J.L.; Yang, X.; Carrier, F.; et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011, 39, 8513–8530. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- McStay, B. Nucleolar organizer regions: Genomic ‘dark matter’ requiring illumination. Genes Dev. 2016, 30, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Azman, M.S.; Alard, E.L.; Dodel, M.; Capraro, F.; Faraway, R.; Dermit, M.; Fan, W.; Chakraborty, A.; Ule, J.; Mardakheh, F.K. An ERK1/2-driven RNA-binding switch in nucleolin drives ribosome biogenesis and pancreatic tumorigenesis downstream of RAS oncogene. Embo J. 2023, 42, e110902. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.C.; Harrington, E.M.; Schumann, S.; Vasconcelos, E.J.R.; Mottram, T.J.; Harper, K.L.; Aspden, J.L.; Whitehouse, A. Kaposi’s sarcoma-associated herpesvirus induces specialised ribosomes to efficiently translate viral lytic mRNAs. Nat. Commun. 2023, 14, 300. [Google Scholar] [CrossRef]
- Atari, N.; Rajan, K.S.; Chikne, V.; Cohen-Chalamish, S.; Doniger, T.; Orbaum, O.; Jacob, A.; Kalt, I.; Michaeli, S.; Sarid, R. Lytic Reactivation of the Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Is Accompanied by Major Nucleolar Alterations. Viruses 2022, 14, 1720. [Google Scholar] [CrossRef]
- Sadagopan, S.; Sharma-Walia, N.; Veettil Mohanan, V.; Bottero, V.; Levine, R.; Vart Richard, J.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus Upregulates Angiogenin during Infection of Human Dermal Microvascular Endothelial Cells, Which Induces 45S rRNA Synthesis, Antiapoptosis, Cell Proliferation, Migration, and Angiogenesis. J. Virol. 2009, 83, 3342–3364. [Google Scholar] [CrossRef]
- Paudel, N.; Sadagopan, S.; Balasubramanian, S.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen and Angiogenin Interact with Common Host Proteins, Including Annexin A2, Which Is Essential for Survival of Latently Infected Cells. J. Virol. 2012, 86, 1589–1607. [Google Scholar] [CrossRef] [PubMed]
- Paudel, N.; Sadagopan, S.; Chakraborty, S.; Sarek, G.; Ojala Päivi, M.; Chandran, B. Kaposi’s Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Interacts with Multifunctional Angiogenin to Utilize Its Antiapoptotic Functions. J. Virol. 2012, 86, 5974–5991. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zareie, A.R.; Verma, S.C. Nucleolin Regulates the Expression of Kaposi’s Sarcoma-Associated Herpesvirus’ Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA. Viruses 2023, 15, 2438. https://doi.org/10.3390/v15122438
Zareie AR, Verma SC. Nucleolin Regulates the Expression of Kaposi’s Sarcoma-Associated Herpesvirus’ Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA. Viruses. 2023; 15(12):2438. https://doi.org/10.3390/v15122438
Chicago/Turabian StyleZareie, Andrew R., and Subhash C. Verma. 2023. "Nucleolin Regulates the Expression of Kaposi’s Sarcoma-Associated Herpesvirus’ Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA" Viruses 15, no. 12: 2438. https://doi.org/10.3390/v15122438
APA StyleZareie, A. R., & Verma, S. C. (2023). Nucleolin Regulates the Expression of Kaposi’s Sarcoma-Associated Herpesvirus’ Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA. Viruses, 15(12), 2438. https://doi.org/10.3390/v15122438




