Protein Arginylation Is Regulated during SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Methods
2.1. Data Sources and Curation
2.2. Bioinformatics Analysis
2.3. Single-Cell RNA-seq Re-Analysis
2.4. Cell Culture
2.5. Viral Infection
2.6. Time-Course Evaluation of Protein Arginylation during Viral Infection
2.7. Chemical Inhibition of Protein Arginylation
2.8. siRNA-Directed Inhibition of ATE1
2.9. Viral Quantification
2.10. Transient Transfection of HEK 293T Cells
2.11. Western Blot
3. Results
3.1. SARS-CoV-2 Infection Modulated the N-Degron Pathway and Increased ATE1 Enzyme Expression
3.2. Increased ATE1 Expression in SARS-CoV-2 Infection Was Correlated with Events Linked to the Endoplasmic Reticulum (ER)
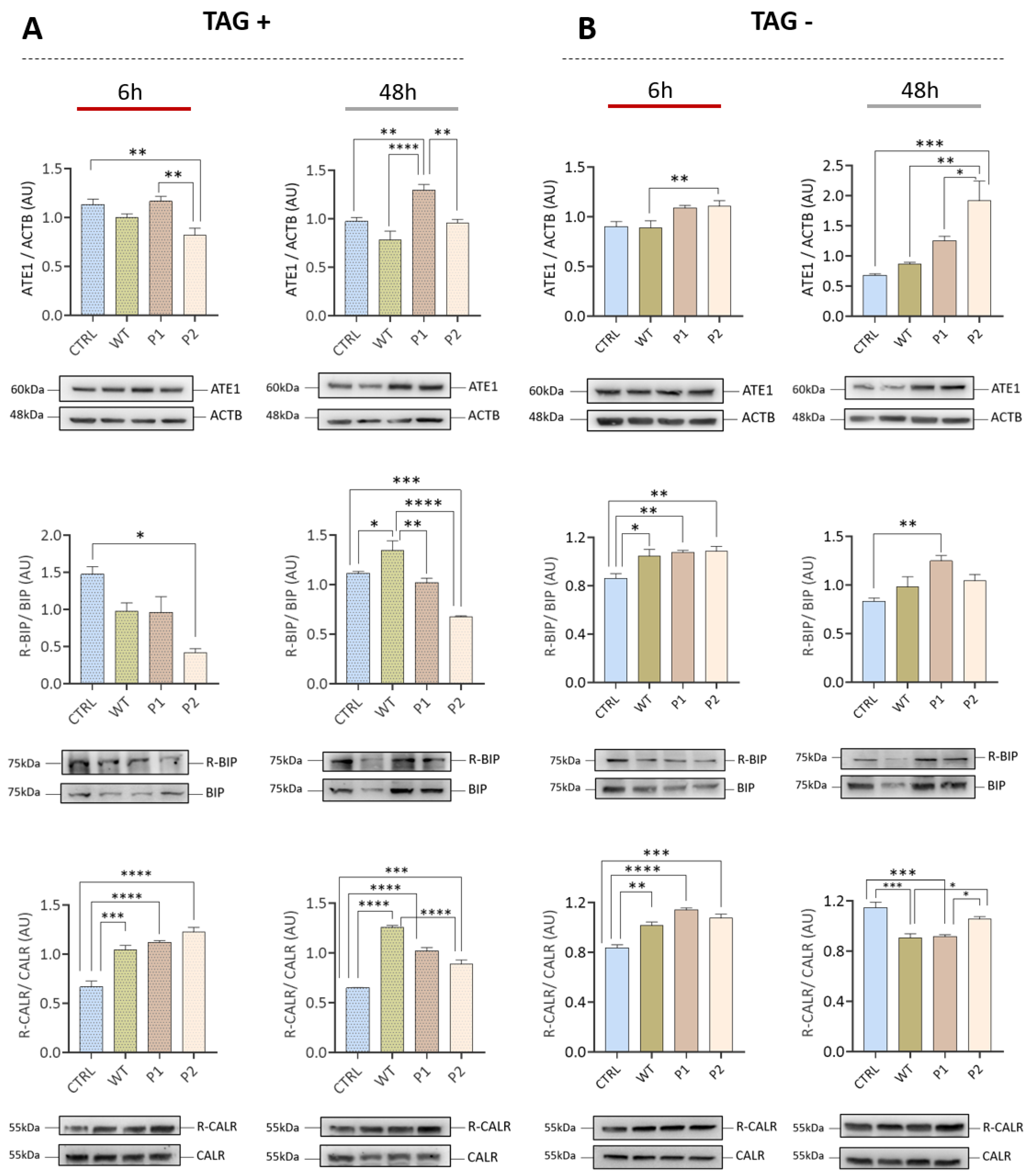
3.3. The Increase in ATE1 Levels Occurs Earlier with the Brazilian Variants P1 and P2 Compared to the Wuhan Variant (WT)
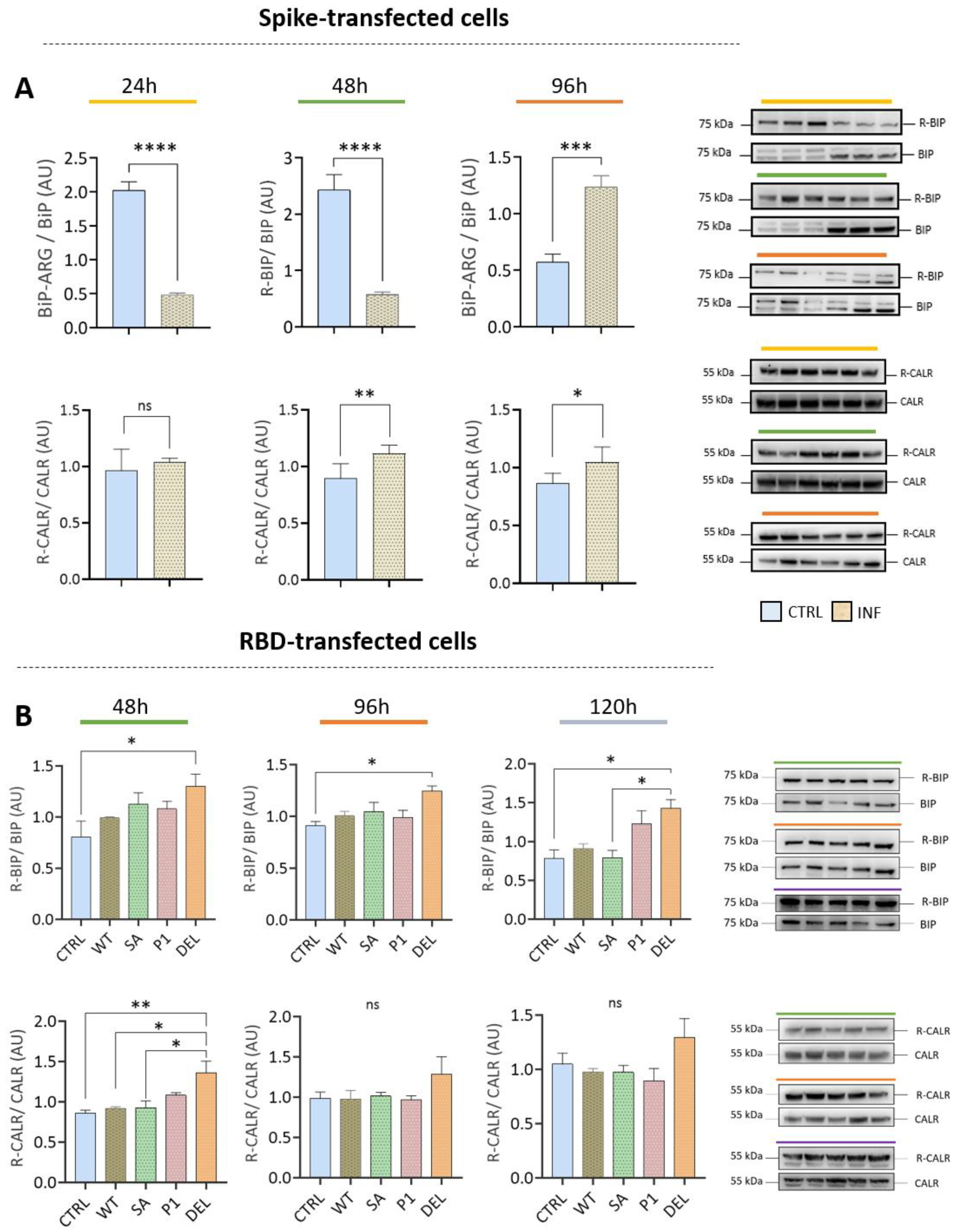
3.4. Transfection with Spike or RBD Highlights the Potential of SARS-CoV-2 to Induce Protein Arginylation
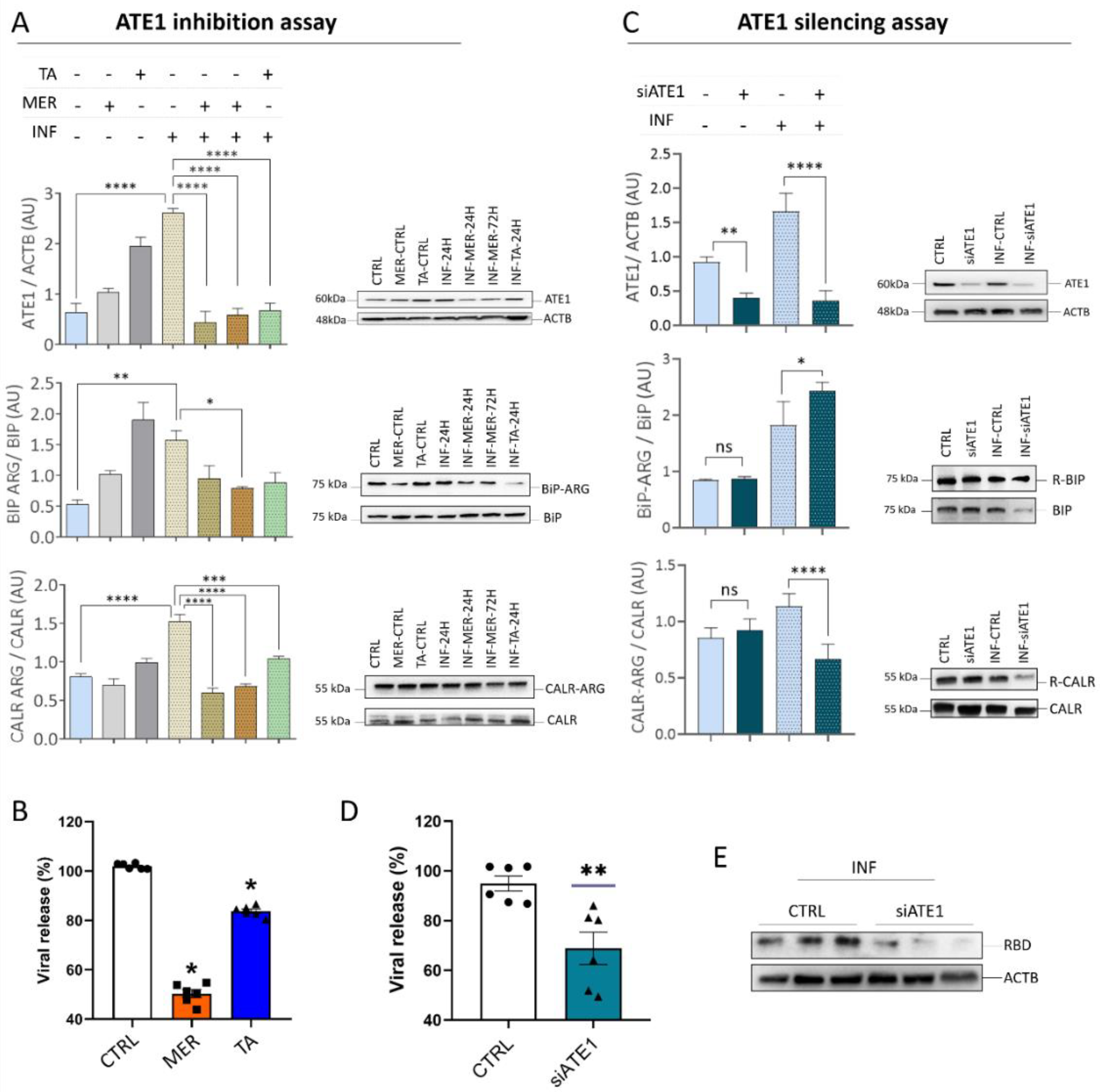
3.5. ATE1 Inhibition and Silencing Reduces SARS-CoV-2 Viral Release in Calu-3 Cells
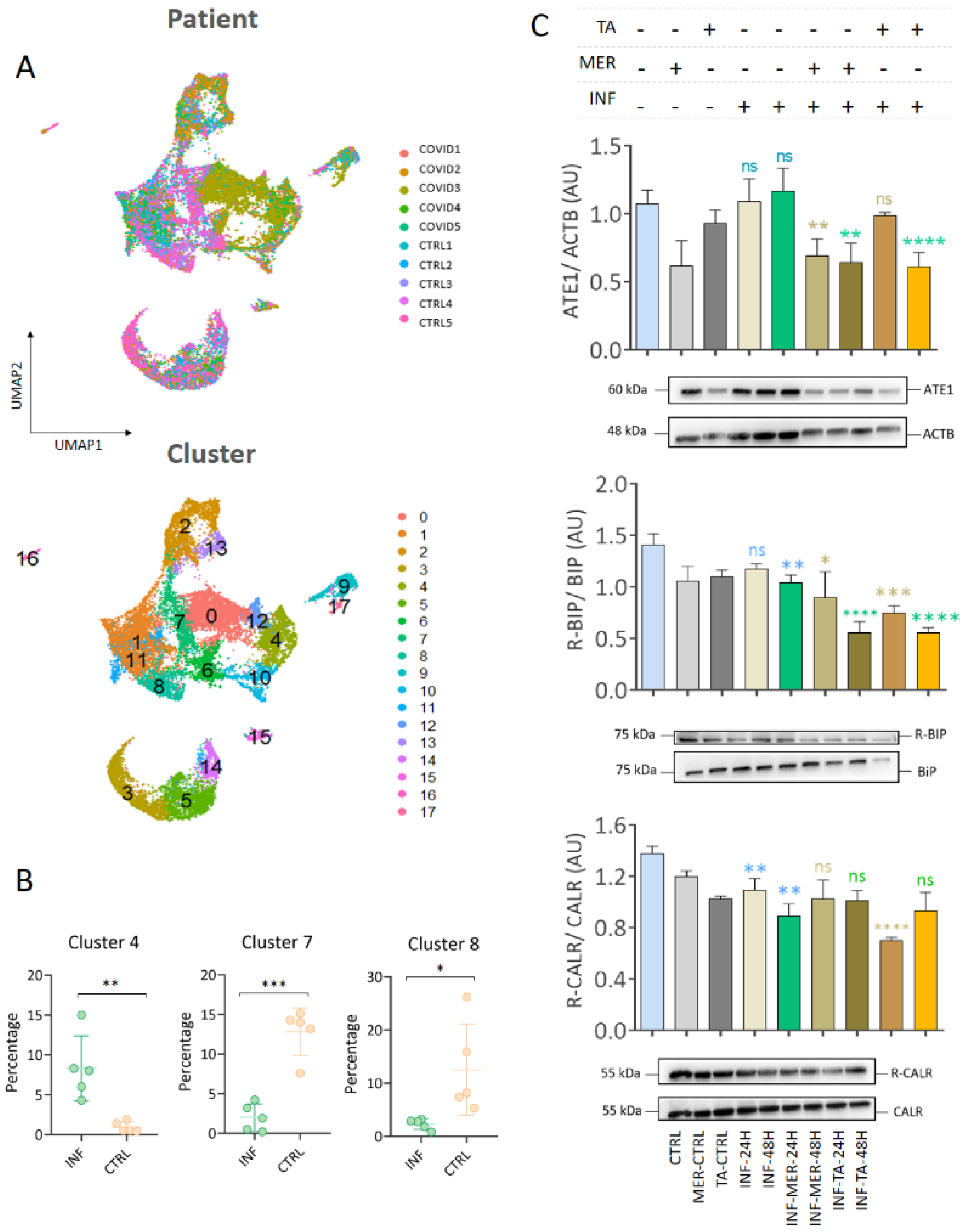
3.6. Single Cell RNA-seq Data Showed That Macrophages and Epithelial Cells Express ATE1
3.7. The N-Degron Pathway Was Regulated in SARS-CoV and MERS-CoV Infections but Not in H1N1 Influenza and Respiratory Syncytial Virus (RSV) Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahalmani, V.M.; Mahendru, D.; Semwal, A.; Kaur, S.; Kaur, H.; Sarma, P.; Prakash, A.; Medhi, B. COVID-19 pandemic: A review based on current evidence. Indian J. Pharmacol. 2020, 52, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A.; et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Gandhi, R.T.; Lynch, J.B.; del Rio, C. Mild or Moderate Covid-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef]
- Esakandari, H.; Nabi-Afjadi, M.; Fakkari-Afjadi, J.; Farahmandian, N.; Miresmaeili, S.-M.; Bahreini, E. A comprehensive review of COVID-19 characteristics. Biol. Proced. Online 2020, 22, 19. [Google Scholar] [CrossRef]
- He, X.; Cheng, X.; Feng, X.; Wan, H.; Chen, S.; Xiong, M. Clinical Symptom Differences between Mild and Severe COVID-19 Patients in China: A Meta-Analysis. Front. Public Health 2021, 8, 561264. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.-S.; Ye, Z.-W.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef]
- Guo, Q.; He, Z. Prediction of the confirmed cases and deaths of global COVID-19 using artificial intelligence. Environ. Sci. Pollut. Res. Int. 2021, 28, 11672–11682. [Google Scholar] [CrossRef]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar] [PubMed]
- Sixto-López, Y.; Correa-Basurto, J.; Bello, M.; Landeros-Rivera, B.; Garzón-Tiznado, J.A.; Montaño, S. Structural insights into SARS-CoV-2 spike protein and its natural mutants found in Mexican population. Sci. Rep. 2021, 11, 4659. [Google Scholar] [CrossRef] [PubMed]
- Hatmal, M.M.; Alshaer, W.; Al-Hatamleh, M.A.I.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive Structural and Molecular Comparison of Spike Proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and Their Interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef]
- Chan, C.-P.; Siu, K.-L.; Chin, K.-T.; Yuen, K.-Y.; Zheng, B.; Jin, D.-Y. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2006, 80, 9279–9287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, T.S.; Huang, M.; Liu, D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res. 2014, 194, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Köseler, A.; Sabirli, R.; Gören, T.; Türkçüer, İ.; Kurt, Ö. Endoplasmic Reticulum Stress Markers in SARS-CoV-2 Infection and Pneumonia: Case-Control Study. In Vivo 2020, 34, 1645–1650. [Google Scholar] [CrossRef]
- Rosa-Fernandes, L.; Lazari, L.C.; da Silva, J.M.; de Morais Gomes, V.; Machado, R.R.G.; dos Santos, A.F.; Araujo, D.B.; Coutinho, J.V.P.; Arini, G.S.; Angeli, C.B.; et al. SARS-CoV-2 activates ER stress and Unfolded protein response. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rashid, F.; Dzakah, E.E.; Wang, H.; Tang, S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021, 296, 198350. [Google Scholar] [CrossRef]
- Balakrishnan, B.; Lai, K. Modulation of SARS-CoV-2 Spike-induced Unfolded Protein Response (UPR) in HEK293T cells by selected small chemical molecules. bioRxiv 2021. [Google Scholar] [CrossRef]
- Aoe, T. Pathological Aspects of COVID-19 as a Conformational Disease and the Use of Pharmacological Chaperones as a Potential Therapeutic Strategy. Front. Pharmacol. 2020, 11, 1095. [Google Scholar] [CrossRef]
- Barabutis, N. Unfolded Protein Response in Acute Respiratory Distress Syndrome. Lung 2019, 197, 827–828. [Google Scholar] [CrossRef] [PubMed]
- Bouchecareilh, M.; Balch, W.E. Proteostasis: A new therapeutic paradigm for pulmonary disease. Proc. Am. Thorac. Soc. 2011, 8, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. JASN 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Robbins, J. Proteasomal and lysosomal protein degradation and heart disease. J. Mol. Cell. Cardiol. 2014, 71, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Ravid, T.; Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008, 9, 679–690. [Google Scholar] [CrossRef] [Green Version]
- Livneh, I.; Kravtsova-Ivantsiv, Y.; Braten, O.; Kwon, Y.T.; Ciechanover, A. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. BioEssays 2017, 39, 1700027. [Google Scholar] [CrossRef]
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-end rule pathway. Annu. Rev. Biochem. 2012, 81, 261–289. [Google Scholar] [CrossRef] [Green Version]
- Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011, 20, 1298–1345. [Google Scholar] [CrossRef] [Green Version]
- Kopitz, J.; Rist, B.; Bohley, P. Post-translational arginylation of ornithine decarboxylase from rat hepatocytes. Biochem. J. 1990, 267, 343–348. [Google Scholar] [CrossRef] [Green Version]
- Eriste, E.; Norberg, A.; Nepomuceno, D.; Kuei, C.; Kamme, F.; Tran, D.-T.; Strupat, K.; Jörnvall, H.; Liu, C.; Lovenberg, T.W.; et al. A novel form of neurotensin post-translationally modified by arginylation. J. Biol. Chem. 2005, 280, 35089–35097. [Google Scholar] [CrossRef]
- Soffer, R.L. Enzymatic arginylation of beta-melanocyte-stimulating hormone and of angiotensin II. J. Biol. Chem. 1975, 250, 2626–2629. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, X.; Leu, N.A.; Sterling, S.; Kurosaka, S.; Fina, M.; Lee, V.M.; Dong, D.W.; Yates, J.R.; Kashina, A. Protein arginylation targets alpha synuclein, facilitates normal brain health, and prevents neurodegeneration. Sci. Rep. 2017, 7, 11323. [Google Scholar] [CrossRef]
- Hu, R.-G.; Wang, H.; Xia, Z.; Varshavsky, A. The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. USA 2008, 105, 76–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha-Molstad, H.; Kwon, Y.T.; Kim, B.Y. Amino-terminal arginylation as a degradation signal for selective autophagy. BMB Rep. 2015, 48, 487–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Han, X.; Wong, C.C.L.; Cheng, H.; Aslanian, A.; Xu, T.; Leavis, P.; Roder, H.; Hedstrom, L.; Yates, J.R.; et al. Arginyltransferase ATE1 catalyzes midchain arginylation of proteins at side chain carboxylates in vivo. Chem. Biol. 2014, 21, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Pejaver, V.R.; Dann, G.P.; Wolf, M.Y.; Kellis, M.; Huang, Y.; Garcia, B.A.; Radivojac, P.; Kashina, A. Target site specificity and in vivo complexity of the mammalian arginylome. Sci. Rep. 2018, 8, 16177. [Google Scholar] [CrossRef] [Green Version]
- Deka, K.; Saha, S. Arginylation: A new regulator of mRNA stability and heat stress response. Cell Death Dis. 2017, 8, e2604. [Google Scholar] [CrossRef]
- Kumar, A.; Birnbaum, M.D.; Patel, D.M.; Morgan, W.M.; Singh, J.; Barrientos, A.; Zhang, F. Posttranslational arginylation enzyme Ate1 affects DNA mutagenesis by regulating stress response. Cell Death Dis. 2016, 7, e2378. [Google Scholar] [CrossRef] [Green Version]
- Deka, K.; Singh, A.; Chakraborty, S.; Mukhopadhyay, R.; Saha, S. Protein arginylation regulates cellular stress response by stabilizing HSP70 and HSP40 transcripts. Cell Death Discov. 2016, 2, 16074. [Google Scholar] [CrossRef]
- Xue, M.; Feng, L. The Role of Unfolded Protein Response in Coronavirus Infection and Its Implications for Drug Design. Front. Microbiol. 2021, 12, 808593. [Google Scholar] [CrossRef] [PubMed]
- Echavarría-Consuegra, L.; Cook, G.M.; Busnadiego, I.; Lefèvre, C.; Keep, S.; Brown, K.; Doyle, N.; Dowgier, G.; Franaszek, K.; Moore, N.A.; et al. Manipulation of the unfolded protein response: A pharmacological strategy against coronavirus infection. PLoS Pathog. 2021, 17, e1009644. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Kashina, A. Posttranslational arginylation as a global biological regulator. Dev. Biol. 2011, 358, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Xia, X.; Yang, X.; Zhang, X.; Liu, Y.; Wu, D.; Fang, Y.; Liu, Y.; Xu, J.; Qiu, Y.; et al. A picorna-like virus suppresses the N-end rule pathway to inhibit apoptosis. eLife 2017, 6, e30590. [Google Scholar] [CrossRef]
- de Oliveira, G.S.; Kawahara, R.; Rosa-Fernandes, L.; Avila, C.C.; Larsen, M.R.; Pereira Alves, J.M.; Palmisano, G. Novel DNA coding regions and protein arginylation reveal unexplored T. cruzi proteome and PTMs. Int. J. Mass Spectrom. 2017, 418, 51–66. [Google Scholar] [CrossRef]
- Boso, G.; Tasaki, T.; Kwon, Y.T.; Somia, N.V. The N-end rule and retroviral infection: No effect on integrase. Virol. J. 2013, 10, 233. [Google Scholar] [CrossRef] [Green Version]
- de Marchi, R.; Sorel, M.; Mooney, B.; Fudal, I.; Goslin, K.; Kwaśniewska, K.; Ryan, P.T.; Pfalz, M.; Kroymann, J.; Pollmann, S.; et al. The N-end rule pathway regulates pathogen responses in plants. Sci. Rep. 2016, 6, 26020. [Google Scholar] [CrossRef] [Green Version]
- Leysath, C.E.; Phillips, D.D.; Crown, D.; Fattah, R.J.; Moayeri, M.; Leppla, S.H. Anthrax Edema Factor Toxicity Is Strongly Mediated by the N-end Rule. PLoS ONE 2013, 8, e74474. [Google Scholar] [CrossRef] [Green Version]
- Schnupf, P.; Zhou, J.; Varshavsky, A.; Portnoy, D.A. Listeriolysin O secreted by Listeria monocytogenes into the host cell cytosol is degraded by the N-end rule pathway. Infect. Immun. 2007, 75, 5135–5147. [Google Scholar] [CrossRef] [Green Version]
- Graciet, E.; Hu, R.-G.; Piatkov, K.; Rhee, J.H.; Schwarz, E.M.; Varshavsky, A. Aminoacyl-transferases and the N-end rule pathway of prokaryotic/eukaryotic specificity in a human pathogen. Proc. Natl. Acad. Sci. USA 2006, 103, 3078–3083. [Google Scholar] [CrossRef]
- Saccon, E.; Chen, X.; Mikaeloff, F.; Rodriguez, J.E.; Szekely, L.; Vinhas, B.S.; Krishnan, S.; Byrareddy, S.N.; Frisan, T.; Végvári, Á.; et al. Cell-type-resolved quantitative proteomics map of interferon response against SARS-CoV-2. iScience 2021, 24, 102420. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Qian, L.; Sun, R.; Huang, B.; Dong, X.; Xiao, Q.; Zhang, Q.; Lu, T.; Yue, L.; Chen, S.; et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell 2021, 184, 775–791.e14. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Cao, R.; Ma, J.; Mou, D.; Zhu, Y.; Li, W.; Lv, L.; Gao, D.; Zhang, S.; Gong, F.; et al. Pathological features of COVID-19-associated lung injury: A preliminary proteomics report based on clinical samples. Signal Transduct. Target. Ther. 2020, 5, 240. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Wu, D.; Ning, W.; Zhang, J.; Shu, T.; Huang, C.; Chen, R.; Huang, M.; Xu, J.; Yang, Q.; et al. Post-mortem tissue proteomics reveals the pathogenesis of multi-organ injuries of COVID-19. Natl. Sci. Rev. 2020, 8, nwab143. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R.; et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef]
- Desai, N.; Neyaz, A.; Szabolcs, A.; Shih, A.R.; Chen, J.H.; Thapar, V.; Nieman, L.T.; Solovyov, A.; Mehta, A.; Lieb, D.J.; et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat. Commun. 2020, 11, 6319. [Google Scholar] [CrossRef]
- Zhuravlev, E.; Sergeeva, M.; Malanin, S.; Amirkhanov, R.; Semenov, D.; Grigoryeva, T.; Komissarov, A.; Stepanov, G. RNA-Seq transcriptome data of human cells infected with influenza A/Puerto Rico/8/1934 (H1N1) virus. Data Brief 2020, 33, 106604. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Fan, W.; Zhang, S.; Li, Y.; Gu, J.; Zhou, J.; Liu, W. Transcriptome Profiling Reveals Differential Effect of Interleukin-17A Upon Influenza Virus Infection in Human Cells. Front. Microbiol. 2019, 10, 2344. [Google Scholar] [CrossRef]
- Krishnamoorthy, P.; Raj, A.S.; Roy, S.; Kumar, N.S.; Kumar, H. Comparative transcriptome analysis of SARS-CoV, MERS-CoV, and SARS-CoV-2 to identify potential pathways for drug repurposing. Comput. Biol. Med. 2021, 128, 104123. [Google Scholar] [CrossRef]
- Ampuero, S.; Andaur, R.; Milano, M.; Moreno, M.; Lizama, L.; Larrañaga, C.; Urzúa, U. Time-course of transcriptome response to respiratory syncytial virus infection in lung epithelium cells. Acta Virol. 2018, 62, 310–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besteman, S.B.; Callaghan, A.; Langedijk, A.C.; Hennus, M.P.; Meyaard, L.; Mokry, M.; Bont, L.J.; Calis, J.J.A. Transcriptome of airway neutrophils reveals an interferon response in life-threatening respiratory syncytial virus infection. Clin. Immunol. 2020, 220, 108593. [Google Scholar] [CrossRef] [PubMed]
- Dave, K.A.; Norris, E.L.; Bukreyev, A.A.; Headlam, M.J.; Buchholz, U.J.; Singh, T.; Collins, P.L.; Gorman, J.J. A comprehensive proteomic view of responses of A549 type II alveolar epithelial cells to human respiratory syncytial virus infection. Mol. Cell. Proteom. MCP 2014, 13, 3250–3269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zecha, J.; Lee, C.-Y.; Bayer, F.P.; Meng, C.; Grass, V.; Zerweck, J.; Schnatbaum, K.; Michler, T.; Pichlmair, A.; Ludwig, C.; et al. Data, Reagents, Assays and Merits of Proteomics for SARS-CoV-2 Research and Testing. Mol. Cell. Proteom. MCP 2020, 19, 1503–1522. [Google Scholar] [CrossRef]
- Seo, T.; Kim, J.; Shin, H.-C.; Kim, J.G.; Ju, S.; Nawale, L.; Han, G.; Lee, H.S.; Bang, G.; Kim, J.Y.; et al. R-catcher, a potent molecular tool to unveil the arginylome. Cell. Mol. Life Sci. 2021, 78, 3725–3741. [Google Scholar] [CrossRef]
- Wong, C.C.L.; Xu, T.; Rai, R.; Bailey, A.O.; Yates, J.R.; Wolf, Y.I.; Zebroski, H.; Kashina, A. Global analysis of posttranslational protein arginylation. PLoS Biol. 2007, 5, e258. [Google Scholar] [CrossRef] [Green Version]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef] [Green Version]
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. In Structural Approaches to Sequence Evolution; Bastolla, U., Porto, M., Roman, H.E., Vendruscolo, M., Eds.; Biological and Medical Physics, Biomedical Engineering; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. ISBN 978-3-540-35305-8. [Google Scholar]
- Breckels, L.M.; Mulvey, C.M.; Lilley, K.S.; Gatto, L. A Bioconductor workflow for processing and analysing spatial proteomics data. F1000Research 2016, 5, 2926. [Google Scholar] [CrossRef] [Green Version]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193-200. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Gatto, F.; Cagliani, R.; Catelani, T.; Guarnieri, D.; Moglianetti, M.; Pompa, P.P.; Bardi, G. PMA-Induced THP-1 Macrophage Differentiation is Not Impaired by Citrate-Coated Platinum Nanoparticles. Nanomaterials 2017, 7, 332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, D.B.; Machado, R.R.G.; Amgarten, D.E.; Malta, F.d.M.; de Araujo, G.G.; Monteiro, C.O.; Candido, E.D.; Soares, C.P.; de Menezes, F.G.; Pires, A.C.C.; et al. SARS-CoV-2 isolation from the first reported patients in Brazil and establishment of a coordinated task network. Mem. Inst. Oswaldo Cruz 2020, 115, e200342. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Lee, J.; Lee, J.H.; Lee, J.W.; Kim, J.H.; Choi, W.H.; Yoo, Y.D.; Cha-Molstad, H.; Kim, B.Y.; Kwon, Y.T.; et al. The arginylation branch of the N-end rule pathway positively regulates cellular autophagic flux and clearance of proteotoxic proteins. Autophagy 2016, 12, 2197–2212. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, S.-Y.; Kim, D.-H.; Park, J.S.; Jeong, S.H.; Choi, Y.W.; Kim, C.-H. Crosstalk between HSPA5 arginylation and sequential ubiquitination leads to AKT degradation through autophagy flux. Autophagy 2021, 17, 961–979. [Google Scholar] [CrossRef] [PubMed]
- López Sambrooks, C.; Carpio, M.A.; Hallak, M.E. Arginylated calreticulin at plasma membrane increases susceptibility of cells to apoptosis. J. Biol. Chem. 2012, 287, 22043–22054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palecanda, A.; Paulauskis, J.; Al-Mutairi, E.; Imrich, A.; Qin, G.; Suzuki, H.; Kodama, T.; Tryggvason, K.; Koziel, H.; Kobzik, L. Role of the scavenger receptor MARCO in alveolar macrophage binding of unopsonized environmental particles. J. Exp. Med. 1999, 189, 1497–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, S.K.; Chu, P.G.; Weiss, L.M. CD163: A specific marker of macrophages in paraffin-embedded tissue samples. Am. J. Clin. Pathol. 2004, 122, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-J.; Gu, Y.; Wang, C.-Z.; Jin, Y.; Wen, X.-M.; Ma, J.-C.; Tang, L.-J.; Mao, Z.-W.; Qian, J.; Lin, J. The M2 macrophage marker CD206: A novel prognostic indicator for acute myeloid leukemia. Oncoimmunology 2020, 9, 1683347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Härtlova, A.; Gierliński, M.; Prescott, A.; Castellvi, J.; Losa, J.H.; Petersen, S.K.; Wenzel, U.A.; Dill, B.D.; Emmerich, C.H.; et al. Triggering MSR1 promotes JNK-mediated inflammation in IL-4-activated macrophages. EMBO J. 2019, 38, e100299. [Google Scholar] [CrossRef] [PubMed]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Yin, G.-Q.; Zeng, H.-X.; Li, Z.-L.; Chen, C.; Zhong, J.-Y.; Xiao, M.-S.; Zeng, Q.; Jiang, W.-H.; Wu, P.-Q.; Zeng, J.-M.; et al. Differential proteomic analysis of children infected with respiratory syncytial virus. Braz. J. Med. Biol. Res. 2021, 54, e9850. [Google Scholar] [CrossRef]
- Heinonen, S.; Velazquez, V.M.; Ye, F.; Mertz, S.; Acero-Bedoya, S.; Smith, B.; Bunsow, E.; Garcia-Mauriño, C.; Oliva, S.; Cohen, D.M.; et al. Immune profiles provide insights into respiratory syncytial virus disease severity in young children. Sci. Transl. Med. 2020, 12, eaaw0268. [Google Scholar] [CrossRef]
- Vedula, P.; Tang, H.-Y.; Speicher, D.W.; Kashina, A. The UPenn COVID Processing Unit Protein Posttranslational Signatures Identified in COVID-19 Patient Plasma. Front. Cell Dev. Biol. 2022, 10, 807149. [Google Scholar] [CrossRef]
- Paladino, L.; Vitale, A.M.; Caruso Bavisotto, C.; Conway de Macario, E.; Cappello, F.; Macario, A.J.L.; Marino Gammazza, A. The Role of Molecular Chaperones in Virus Infection and Implications for Understanding and Treating COVID-19. J. Clin. Med. 2020, 9, 3518. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elshahat, M.E.; Elfiky, A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Cha-Molstad, H.; Sung, K.S.; Hwang, J.; Kim, K.A.; Yu, J.E.; Yoo, Y.D.; Jang, J.M.; Han, D.H.; Molstad, M.; Kim, J.G.; et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 2015, 17, 917–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nauseef, W.M.; McCormick, S.J.; Clark, R.A. Calreticulin functions as a molecular chaperone in the biosynthesis of myeloperoxidase. J. Biol. Chem. 1995, 270, 4741–4747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salati, S.; Genovese, E.; Carretta, C.; Zini, R.; Bartalucci, N.; Prudente, Z.; Pennucci, V.; Ruberti, S.; Rossi, C.; Rontauroli, S.; et al. Calreticulin Ins5 and Del52 mutations impair unfolded protein and oxidative stress responses in K562 cells expressing CALR mutants. Sci. Rep. 2019, 9, 10558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinari, M.; Eriksson, K.K.; Calanca, V.; Galli, C.; Cresswell, P.; Michalak, M.; Helenius, A. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol. Cell 2004, 13, 125–135. [Google Scholar] [CrossRef]
- Choi, J.-A.; Song, C.-H. Insights Into the Role of Endoplasmic Reticulum Stress in Infectious Diseases. Front. Immunol. 2020, 10, 3147. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Xia, H.; Rajsbaum, R.; Xia, X.; Wang, H.; Shi, P.-Y. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell. Mol. Immunol. 2021, 18, 746–748. [Google Scholar] [CrossRef]
- Haglund, C.M.; Welch, M.D. Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 2011, 195, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.; Zhang, Y.; Lin, Z.; Shi, K.; Jiu, Y. Cytoskeleton—A crucial key in host cell for coronavirus infection. J. Mol. Cell Biol. 2021, 12, 968–979. [Google Scholar] [CrossRef]
- Costa, L.B.; Perez, L.G.; Palmeira, V.A.; Macedo e Cordeiro, T.; Ribeiro, V.T.; Lanza, K.; Simões e Silva, A.C. Insights on SARS-CoV-2 Molecular Interactions With the Renin-Angiotensin System. Front. Cell Dev. Biol. 2020, 8, 559841. [Google Scholar] [CrossRef] [PubMed]
- Park, B.K.; Kim, D.; Park, S.; Maharjan, S.; Kim, J.; Choi, J.-K.; Akauliya, M.; Lee, Y.; Kwon, H.-J. Differential Signaling and Virus Production in Calu-3 Cells and Vero Cells upon SARS-CoV-2 Infection. Biomol. Ther. 2021, 29, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Karakozova, M.; Kozak, M.; Wong, C.C.L.; Bailey, A.O.; Yates, J.R.; Mogilner, A.; Zebroski, H.; Kashina, A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 2006, 313, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Mundia, M.M.; Zhang, F.; Demers, R.W.; Korobova, F.; Svitkina, T.; Perieteanu, A.A.; Dawson, J.F.; Kashina, A. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol. Biol. Cell 2010, 21, 1350–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Kashina, A. Quantification of intracellular N-terminal β-actin arginylation. Sci. Rep. 2019, 9, 16669. [Google Scholar] [CrossRef]
- Drazic, A.; Timmerman, E.; Kajan, U.; Marie, M.; Varland, S.; Impens, F.; Gevaert, K.; Arnesen, T. The Final Maturation State of β-actin Involves N-terminal Acetylation by NAA80, not N-terminal Arginylation by ATE1. J. Mol. Biol. 2022, 434, 167397. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Boumaza, A.; Gay, L.; Mezouar, S.; Diallo, A.B.; Michel, M.; Desnues, B.; Raoult, D.; Scola, B.L.; Halfon, P.; Vitte, J.; et al. Monocytes and macrophages, targets of SARS-CoV-2: The clue for Covid-19 immunoparalysis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kim, J.; Hong, S.P.; Choi, S.Y.; Yang, M.J.; Ju, Y.S.; Kim, Y.T.; Kim, H.M.; Rahman, M.T.; Chung, M.K.; et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Investig. 2021, 131, e148517. [Google Scholar] [CrossRef]
- Wang, S.-C.; Chen, Y.; Wang, Y.-C.; Wang, W.-J.; Yang, C.-S.; Tsai, C.-L.; Hou, M.-H.; Chen, H.-F.; Shen, Y.-C.; Hung, M.-C. Tannic acid suppresses SARS-CoV-2 as a dual inhibitor of the viral main protease and the cellular TMPRSS2 protease. Am. J. Cancer Res. 2020, 10, 4538–4546. [Google Scholar]
- Salgado-Benvindo, C.; Thaler, M.; Tas, A.; Ogando, N.S.; Bredenbeek, P.J.; Ninaber, D.K.; Wang, Y.; Hiemstra, P.S.; Snijder, E.J.; van Hemert, M.J. Suramin Inhibits SARS-CoV-2 Infection in Cell Culture by Interfering with Early Steps of the Replication Cycle. Antimicrob. Agents Chemother. 2020, 64, e00900-20. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Wang, J.; Buckley, B.; Wang, Q.; Lilly, B.; Chernov, M.; Kashina, A. Small molecule inhibitors of arginyltransferase regulate arginylation-dependent protein degradation, cell motility, and angiogenesis. Biochem. Pharmacol. 2012, 83, 866–873. [Google Scholar] [CrossRef] [PubMed]




| Antibody | Secondary Antibody | Dilution | Reference | Type | Company |
|---|---|---|---|---|---|
| Anti-ATE1 | Goat Anti-Rat | 1:1000 | MABS436 | Monoclonal | Merck Millipore |
| Anti-R-BIP | Goat Anti-Rabbit | 1:1000 | ABS2103 | Polyclonal | Merck Millipore |
| Anti-R-PDI | Goat Anti-Rabbit | 1:1000 | ABS1655 | Polyclonal | Merck Millipore |
| Anti-R-CALR | Goat Anti-Rabbit | 1:1000 | ABS1671 | Polyclonal | Merck Millipore |
| Anti-R-ACTB | Goat Anti-Rabbit | 1:1000 | ABT264 | Polyclonal | Merck Millipore |
| Anti-BIP | Goat Anti-Rabbit | 1:1000 | #3183 | Polyclonal | Cell Signaling |
| Anti-PDI | Goat Anti-Mouse | 1:1000 | #MA3-019 | Monoclonal | Sigma-Aldrich |
| Anti-CALR | Goat Anti-Rabbit | 1:1000 | ab2907 | Polyclonal | Abcam |
| Anti-ACTB | Goat Anti-Mouse | 1:10,000 | #A2228 | Monoclonal | Sigma-Aldrich |
| Anti-GAPDH | Goat Anti-Mouse | 1:500 | sc-137179 | Monoclonal | Santa Cruz Biotechnology |
| Anti-UB | Goat Anti-Mouse | 1:1000 | sc-8017 | Monoclonal | Santa Cruz Biotechnology |
| Anti-p62 | Goat Anti-Rabbit | 1:1000 | g # PA5-27247 | Polyclonal | ThermoFisher |
| Anti-LC3BII | Goat Anti-Rabbit | 1:1000 | # PA1-16930 | Polyclonal | ThermoFisher |
| Goat Anti-Rat | - | 1:1000 | BA-9400 | Polyclonal | Vector Laboratories |
| Goat Anti-Rabbit | - | 1:4000 | ab6721 | Polyclonal | Abcam |
| Goat Anti-Mouse | - | 1:4000 | ab6789 | Polyclonal | Abcam |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macedo-da-Silva, J.; Rosa-Fernandes, L.; Gomes, V.d.M.; Santiago, V.F.; Santos, D.M.; Molnar, C.M.S.; Barboza, B.R.; de Souza, E.E.; Marques, R.F.; Boscardin, S.B.; et al. Protein Arginylation Is Regulated during SARS-CoV-2 Infection. Viruses 2023, 15, 290. https://doi.org/10.3390/v15020290
Macedo-da-Silva J, Rosa-Fernandes L, Gomes VdM, Santiago VF, Santos DM, Molnar CMS, Barboza BR, de Souza EE, Marques RF, Boscardin SB, et al. Protein Arginylation Is Regulated during SARS-CoV-2 Infection. Viruses. 2023; 15(2):290. https://doi.org/10.3390/v15020290
Chicago/Turabian StyleMacedo-da-Silva, Janaina, Livia Rosa-Fernandes, Vinicius de Morais Gomes, Veronica Feijoli Santiago, Deivid Martins Santos, Catarina Maria Stanischesk Molnar, Bruno Rafael Barboza, Edmarcia Elisa de Souza, Rodolfo Ferreira Marques, Silvia Beatriz Boscardin, and et al. 2023. "Protein Arginylation Is Regulated during SARS-CoV-2 Infection" Viruses 15, no. 2: 290. https://doi.org/10.3390/v15020290








