Comparison of the Diagnostic Performance of Deep Learning Algorithms for Reducing the Time Required for COVID-19 RT–PCR Testing
Abstract
:1. Introduction
Related Works
2. Materials and Methods
2.1. Study Participants
2.2. Materials
2.3. Data Description
2.4. Development of the DL Model
2.5. Training and Test Datasets
2.6. Outcomes
2.7. Statistical Analysis
3. Results
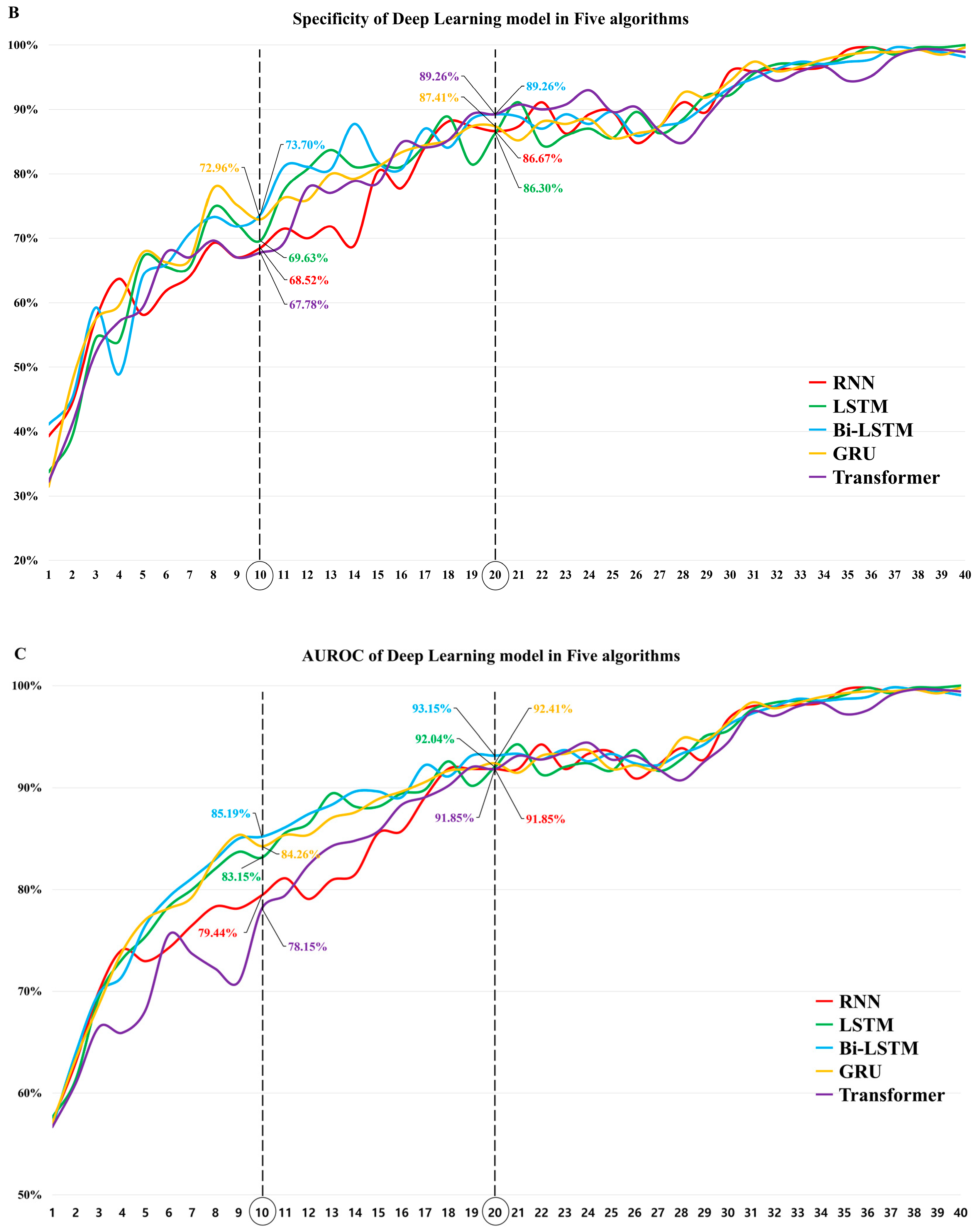
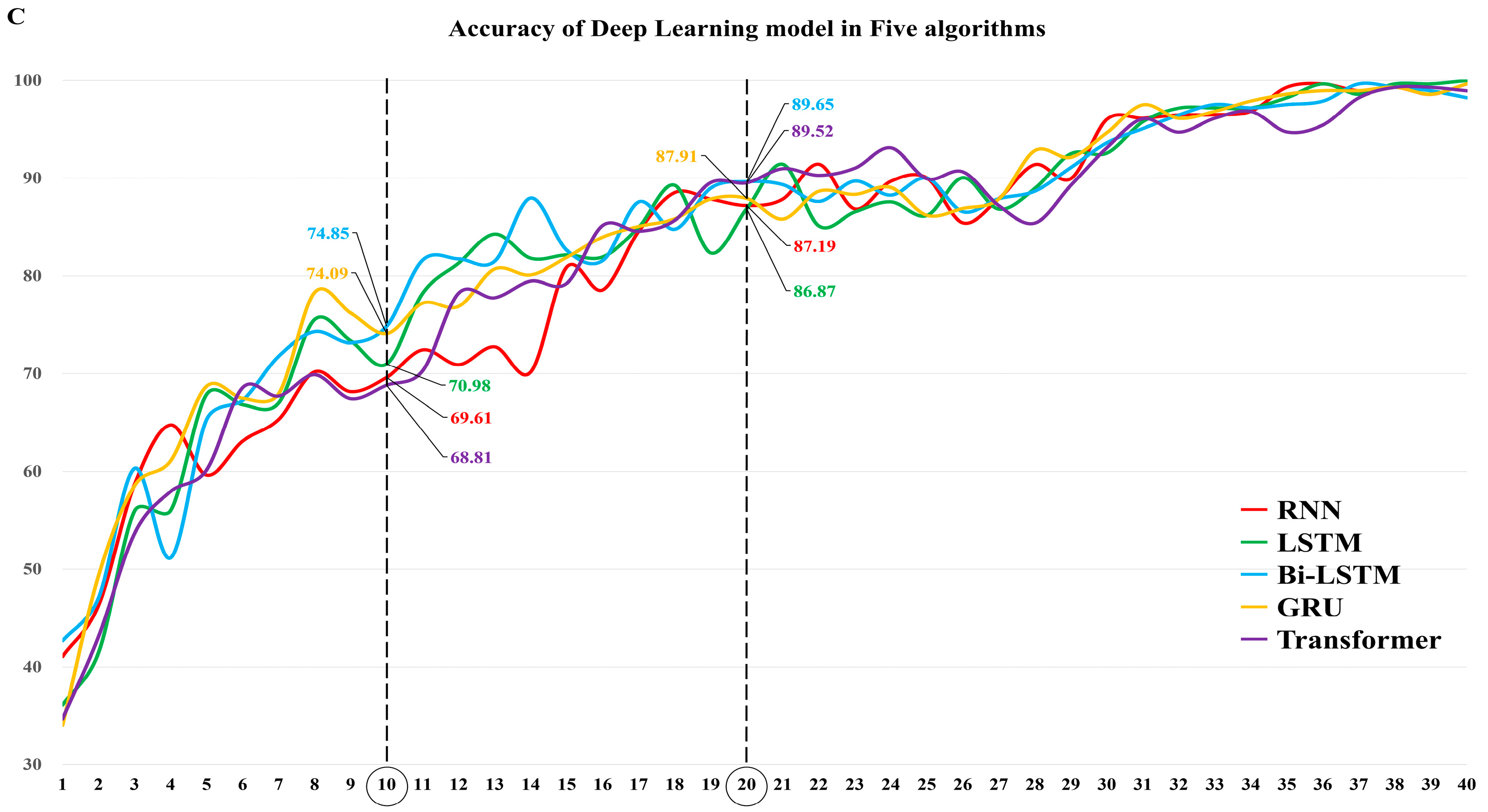
3.1. The Diagnostic Performance of Each DL Algorithm
3.2. The Effect of the Prevalence on the Diagnostic Performance of Each DL Algorithm
3.3. Comparison of the Diagnostic Performance of the Algorithms in Models 10 and 20
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Amer, R.M.; Samir, M.; Gaber, O.A.; El-Deeb, N.A.; Abdelmoaty, A.A.; Ahmed, A.A.; Samy, W.; Atta, A.H.; Walaa, M.; Anis, R.H. Diagnostic performance of rapid antigen test for COVID-19 and the effect of viral load, sampling time, subject’s clinical and laboratory parameters on test accuracy. J. Infect. Public Health 2021, 14, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
- Jegerlehner, S.; Suter-Riniker, F.; Jent, P.; Bittel, P.; Nagler, M. Diagnostic accuracy of a SARS-CoV-2 rapid antigen test in real-life clinical settings. Int. J. Infect. Dis. 2021, 109, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Holzner, C.; Pabst, D.; Anastasiou, O.E.; Dittmer, U.; Manegold, R.K.; Risse, J.; Fistera, D.; Kill, C.; Falk, M. SARS-CoV-2 rapid antigen test: Fast-safe or dangerous? An analysis in the emergency department of an university hospital. J. Med. Virol. 2021, 93, 5323–5327. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Maurici, M.; Pieri, M.; Andreoni, M.; Bernardini, S. Performance of a rapid antigen test in the diagnosis of SARS-CoV-2 infection. J. Med. Virol. 2021, 93, 2988–2991. [Google Scholar] [CrossRef]
- Onsongo, S.N.; Otieno, K.; van Duijn, S.; Adams, E.; Omollo, M.; Odero, I.A.; K’Oloo, A.; Houben, N.; Milimo, E.; Aroka, R.; et al. Performance of a rapid antigen test for SARS-CoV-2 in Kenya. Diagn. Microbiol. Infect. Dis. 2022, 102, 115591. [Google Scholar] [CrossRef]
- Turcato, G.; Zaboli, A.; Pfeifer, N.; Sibilio, S.; Tezza, G.; Bonora, A.; Ciccariello, L.; Ausserhofer, D. Rapid antigen test to identify COVID-19 infected patients with and without symptoms admitted to the Emergency Department. Am. J. Emerg. Med. 2022, 51, 92–97. [Google Scholar] [CrossRef]
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, Y.S.; Lee, D.I.; Jeong, S.; Kang, G.H.; Jang, Y.S.; Kim, W.; Choi, H.Y.; Kim, J.G.; Choi, S.H. The application of a deep learning system developed to reduce the time for RT–PCR in COVID-19 detection. Sci. Rep. 2022, 12, 1234. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning internal representations by error propagation. Calif. Univ. San Diego La Jolla Inst. Cogn. Sci. 1985, 1–34. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.; Van Merriënboer, B.; Bahdanau, D.; Bengio, Y. On the properties of neural machine translation: Encoder-decoder approaches. arXiv 2015, arXiv:1409.1259. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- The Johns Hopkins University. COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE). Available online: https://github.com/CSSEGISandData/COVID-19 (accessed on 25 February 2022).
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression; John Wiley and Sons: New York, NY, USA, 2000. [Google Scholar]
- Kim, J.; Hwang, I.C. Drawing guidelines for receiver operating characteristic curve in preparation of manuscripts. J. Korean Med. Sci. 2020, 35, e171. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hong, H.; Yoon, S.H. Diagnostic performance of CT and reverse transcriptase polymerase chain reaction for coronavirus disease 2019: A meta-analysis. Radiology 2020, 296, E145–E155. [Google Scholar] [CrossRef]
- Hayer, J.; Kasapic, D.; Zemmrich, C. Real-world clinical performance of commercial SARS-CoV-2 rapid antigen tests in suspected COVID-19: A systematic meta-analysis of available data as of November 20, 2020. Int. J. Infect. Dis. 2021, 108, 592–602. [Google Scholar] [CrossRef]
- Riccò, M.; Ferraro, P.; Gualerzi, G.; Ranzieri, S.; Henry, B.M.; Said, Y.B.; Pyatigorskaya, N.V.; Nevolina, E.; Wu, J.; Bragazzi, N.L.; et al. Point-of-care diagnostic tests for detecting SARS-CoV-2 antibodies: A systematic review and meta-analysis of real-world data. J. Clin. Med. 2020, 9, 1515. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.C.; Diao, K.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef]
- Jin, C.; Chen, W.; Cao, Y.; Xu, Z.; Tan, Z.; Zhang, X.; Deng, L.; Zheng, C.; Zhou, J.; Shi, H.; et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat. Commun. 2020, 11, 5088. [Google Scholar] [CrossRef]
- Javor, D.; Kaplan, H.; Kaplan, A.; Puchner, S.B.; Krestan, C.; Baltzer, P. Deep learning analysis provides accurate COVID-19 diagnosis on chest computed tomography. Eur. J. Radiol. 2020, 133, 109402. [Google Scholar] [CrossRef] [PubMed]
- Serte, S.; Demirel, H. Deep learning for diagnosis of COVID-19 using 3D CT scans. Comput. Biol. Med. 2021, 132, 104306. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.; Esfahanian, P.; Movahed, S.M.S.; Gorgin, S.; Rahmati, D.; Abedini, A.; Nadji, S.A.; Haseli, S.; Bakhshayesh Karam, M.; Kiani, A.; et al. ai-corona: Radiologist-assistant deep learning framework for COVID-19 diagnosis in chest CT scans. PLoS ONE 2021, 16, e0250952. [Google Scholar]
- Shah, V.; Keniya, R.; Shridharani, A.; Punjabi, M.; Shah, J.; Mehendale, N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021, 28, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, L.; Jin, R.; Liang, L.; Hu, Z.; Tao, L.; Han, Y.; Feng, W.; Zhou, D.; Li, W.; et al. Texture feature-based machine learning classifier could assist in the diagnosis of COVID-19. Eur. J. Radiol. 2021, 137, 109602. [Google Scholar] [CrossRef]
- Wang, S.; Zha, Y.; Li, W.; Wu, Q.; Li, X.; Niu, M.; Wang, M.; Qiu, X.; Li, H.; Yu, H.; et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020, 56, 2000775. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy. Radiology 2020, 296, E65–E71. [Google Scholar] [CrossRef] [PubMed]
- Fontanellaz, M.; Ebner, L.; Huber, A.; Peters, A.; Löbelenz, L.; Hourscht, C.; Klaus, J.; Munz, J.; Ruder, T.; Drakopoulos, D.; et al. A deep-learning diagnostic support system for the detection of COVID-19 using chest radiographs: A multireader validation study. Invest. Radiol. 2021, 56, 348–356. [Google Scholar] [CrossRef]
- Wang, D.; Mo, J.; Zhou, G.; Xu, L.; Liu, Y. An efficient mixture of deep and machine learning models for COVID-19 diagnosis in chest X-ray images. PLoS One 2020, 15, e0242535. [Google Scholar] [CrossRef]
- Carlile, M.; Hurt, B.; Hsiao, A.; Hogarth, M.; Longhurst, C.A.; Dameff, C. Deployment of artificial intelligence for radiographic diagnosis of COVID-19 pneumonia in the emergency department. J. Am. Coll. Emerg. Phys. Open 2020, 1, 1459–1464. [Google Scholar] [CrossRef]
- Zhang, R.; Tie, X.; Qi, Z.; Bevins, N.B.; Zhang, C.; Griner, D.; Song, T.K.; Nadig, J.D.; Schiebler, M.L.; Garrett, J.W.; et al. Diagnosis of coronavirus disease 2019 pneumonia by using chest radiography: Value of artificial intelligence. Radiology 2021, 298, E88–E97. [Google Scholar] [CrossRef]
- Wang, G.; Liu, X.; Shen, J.; Wang, C.; Li, Z.; Ye, L.; Wu, X.; Chen, T.; Wang, K.; Zhang, X.; et al. A deep-learning pipeline for the diagnosis and discrimination of viral, nonviral and COVID-19 pneumonia from chest X-ray images. Nat. Biomed. Eng. 2021, 5, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Khuzani, A.Z.; Heidari, M.; Shariati, S.A. COVID-Classifier: An automated machine learning model to assist in the diagnosis of COVID-19 infection in chest X-ray images. Sci. Rep. 2021, 11, 9887. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, I.; Ippolito, D.; Interlenghi, M.; Monti, C.B.; Salvatore, C.; Schiaffino, S.; Polidori, A.; Gandola, D.; Messa, C.; Sardanelli, F. Machine learning applied on chest X-ray can aid in the diagnosis of COVID-19: A first experience from Lombardy, Italy. Eur. Radiol. Exp. 2021, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.A.; Castro, G.Z.; Oliveira, B.A.S.; Ferreira, L.A.; Ramírez, J.A.; Silva, R.; Guimarães, F.G. Explaining machine learning based diagnosis of COVID-19 from routine blood tests with decision trees and criteria graphs. Comput. Biol. Med. 2021, 132, 104335. [Google Scholar] [CrossRef]
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef]
- Goodman-Meza, D.; Rudas, A.; Chiang, J.N.; Adamson, P.C.; Ebinger, J.; Sun, N.; Botting, P.; Fulcher, J.A.; Saab, F.G.; Brook, R.; et al. A machine learning algorithm to increase COVID-19 inpatient diagnostic capacity. PLoS ONE 2020, 15, e0239474. [Google Scholar] [CrossRef]
- Laghi, A. Cautions about radiologic diagnosis of COVID-19 infection driven by artificial intelligence. Lancet Digit. Health 2020, 2, e225. [Google Scholar] [CrossRef]






| Hyper-Parameters | Optimal Value | Experimental Categories | |
|---|---|---|---|
| Common | Loss function | Cross-entropy | - |
| Optimization algorithm | Adam | SGD, Adam, momentum | |
| Learning rate | 1 × 10−3 | 1 × 10−2–1 × 10−6 | |
| Batch size | 128 | 32–128 | |
| RNN-based approach(RNN, LSTM, Bi-LSTM, GRU) | No. of hidden units | 128 | 32–256 |
| No. of hidden units in dense layer | 32 | 32–128 | |
| Dropout rate | 0.3 | 0.3–0.6 | |
| Transformer | No. of heads | 4 | 2–6 |
| No. of transformer layers | 4 | 2–4 | |
| No. of hidden units in multi-head self-attention | 256 | 64–512 | |
| No. of hidden units in dense layer | 128 | 32–128 | |
| Dropout rate | 0.25 | 0.2–0.6 | |
| Model 10 | AUROC (95% CI) | Pairwise Comparison | p Value | Pairwise Comparison | p Value |
|---|---|---|---|---|---|
| RNN | 79.44 (76.12–82.24) | RNN vs. LSTM | 0.0036 | LSTM vs. GRU | 0.2372 |
| LSTM | 83.15 (80.25–85.92) | RNN vs. Bi-LSTM | <0.0001 | LSTM vs. Transformer | 0.0005 |
| Bi-LSTM | 85.19 (82.22–88.12) | RNN vs. GRU | 0.0003 | Bi-LSTM vs. GRU | 0.2255 |
| GRU | 84.26 (81.56–87.12) | RNN vs. Transformer | 0.2626 | Bi-LSTM vs. Transformer | <0.0001 |
| Transformer | 78.15 (74.88–81.70) | LSTM vs. Bi-LSTM | 0.0154 | GRU vs. Transformer | <0.0001 |
| Model 20 | AUROC (95% CI) | Pairwise Comparison | p Value | Pairwise Comparison | p Value |
|---|---|---|---|---|---|
| RNN | 91.85 (89.68–94.04) | RNN vs. LSTM | 0.8275 | LSTM vs. GRU | 0.5638 |
| LSTM | 92.04 (89.86–94.19) | RNN vs. Bi-LSTM | 0.1257 | LSTM vs. transformer | 0.8565 |
| Bi-LSTM | 93.15 (91.04–95.03) | RNN vs. GRU | 0.5322 | Bi-LSTM vs. GRU | 0.3708 |
| GRU | 92.41 (90.30–94.48) | RNN vs. transformer | 1.0000 | Bi-LSTM vs. transformer | 0.1931 |
| Transformer | 91.85 (89.50–94.00) | LSTM vs. Bi-LSTM | 0.1313 | GRU vs. transformer | 0.6004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.; Kim, Y.-S.; Lee, D.I.; Jeong, S.; Kang, G.H.; Jang, Y.S.; Kim, W.; Choi, H.Y.; Kim, J.G. Comparison of the Diagnostic Performance of Deep Learning Algorithms for Reducing the Time Required for COVID-19 RT–PCR Testing. Viruses 2023, 15, 304. https://doi.org/10.3390/v15020304
Lee Y, Kim Y-S, Lee DI, Jeong S, Kang GH, Jang YS, Kim W, Choi HY, Kim JG. Comparison of the Diagnostic Performance of Deep Learning Algorithms for Reducing the Time Required for COVID-19 RT–PCR Testing. Viruses. 2023; 15(2):304. https://doi.org/10.3390/v15020304
Chicago/Turabian StyleLee, Yoonje, Yu-Seop Kim, Da In Lee, Seri Jeong, Gu Hyun Kang, Yong Soo Jang, Wonhee Kim, Hyun Young Choi, and Jae Guk Kim. 2023. "Comparison of the Diagnostic Performance of Deep Learning Algorithms for Reducing the Time Required for COVID-19 RT–PCR Testing" Viruses 15, no. 2: 304. https://doi.org/10.3390/v15020304





