Three Phages from a Boreal Lake during Ice Cover Infecting Xylophilus, Caulobacter, and Polaromonas Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and 16S rRNA Gene Sequencing of Bacteria
2.2. Phage Sampling and Enrichment
2.3. Phage Culturing and Host Range Analysis
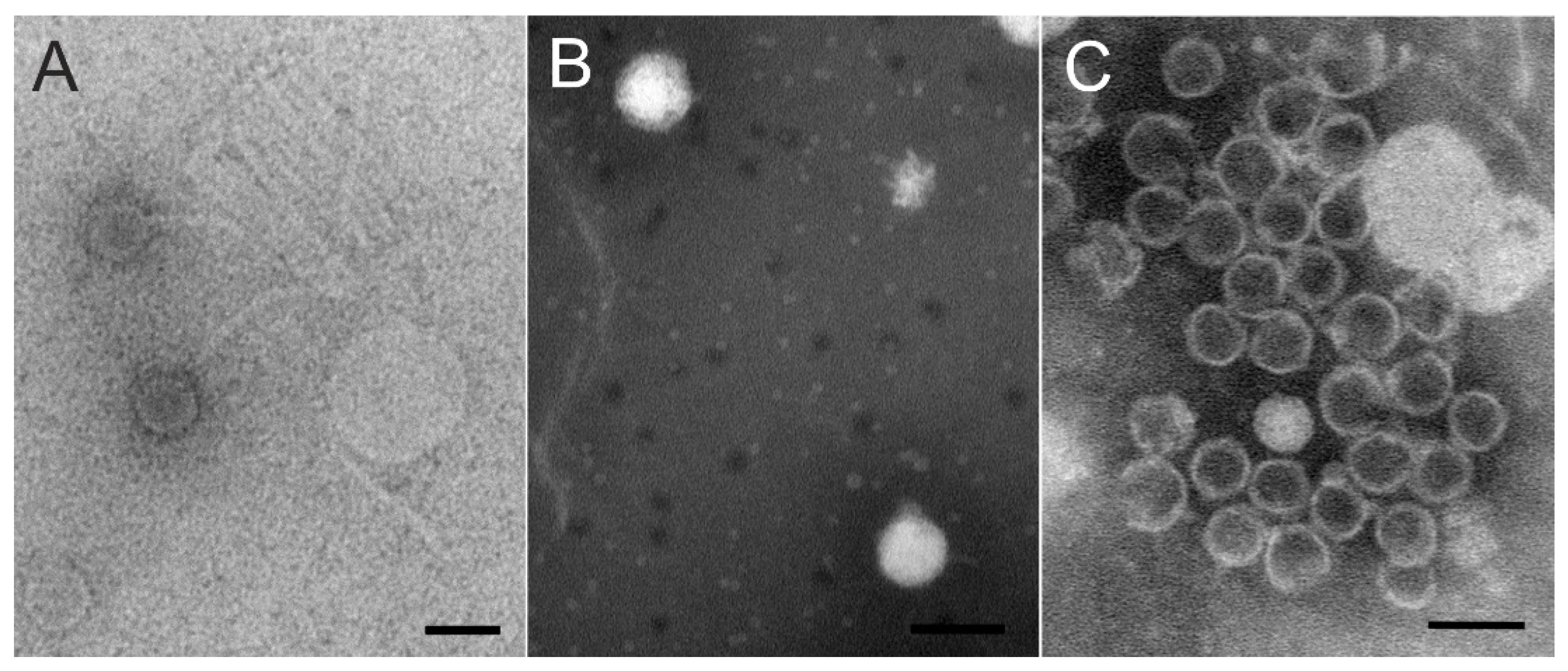
2.4. Phage Purification and Transmission Electron Microscopy
2.5. Phage Genome Sequencing and Analysis
3. Results
3.1. Bacterial Species Isolated under Ice Cover
3.2. Three Unique Tailed Double-Stranded DNA Phages Were Isolated from a Boreal Lake Water Sample during Ice Cover
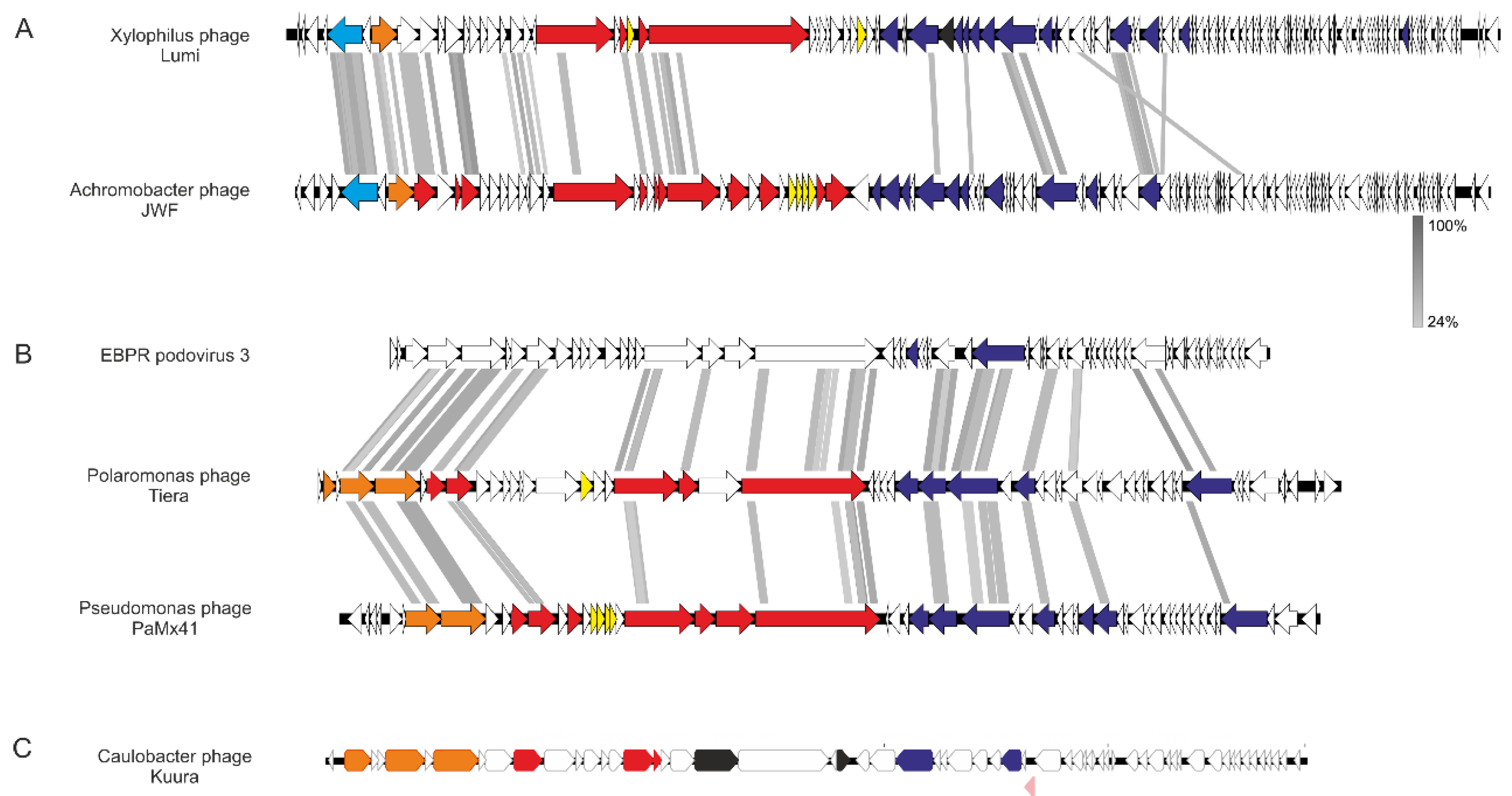
3.2.1. Phage Lumi Infecting Xylophilus sp. B14 Shows Relatedness with Achromobacter Phage JWF
3.2.2. Two Podoviruses Were Isolated, Phage Kuura Infecting Caulobacter sp. B15 and Phage Tiera Infecting Polaromonas sp. B16
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilhelm, S.W.; Suttle, C.A. Viruses and nutrient cycles in the sea. Bioscience 1999, 49, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Braga, L.P.P.; Orland, C.; Emilson, E.J.S.; Fitch, A.A.; Osterholz, H.; Dittmar, T.; Basiliko, N.; Mykytczuk, N.C.S.; Tanentzap, A.J. Viruses direct carbon cycling in lake sediments under global change. Proc. Natl. Acad. Sci. USA 2022, 119, e2202261119. [Google Scholar] [CrossRef] [PubMed]
- Butler, T.M.; Wilhelm, A.-C.; Dwyer, A.C.; Webb, P.N.; Baldwin, A.L.; Techtmann, S.M. Microbial community dynamics during lake ice freezing. Sci. Rep. 2019, 9, 6231. [Google Scholar] [CrossRef] [Green Version]
- Bižić-Ionescu, M.; Amann, R.; Grossart, H.-P. Massive regime shifts and high activity of heterotrophic bacteria in an ice-covered lake. PLoS ONE 2014, 9, e113611. [Google Scholar] [CrossRef]
- Wetzel, R.G. 7—Bacterioplankton. In Limnology; Wetzel, R.G., Ed.; Academic Press: Cambridge, MA, USA, 2001; pp. 489–525. ISBN 9780127447605. [Google Scholar]
- Laanto, E.; Mäntynen, S.; De Colibus, L.; Marjakangas, J.; Gillum, A.; Stuart, D.I.; Ravantti, J.J.; Huiskonen, J.T.; Sundberg, L.-R. Virus found in a boreal lake links ssDNA and dsDNA viruses. Proc. Natl. Acad. Sci. USA 2017, 114, 8378–8383. [Google Scholar] [CrossRef] [Green Version]
- Moon, K.; Jeon, J.H.; Kang, I.; Park, K.S.; Lee, K.; Cha, C.-J.; Lee, S.H.; Cho, J.-C. Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 2020, 8, 75. [Google Scholar] [CrossRef]
- Aguirre de Cárcer, D.; López-Bueno, A.; Pearce, D.A.; Alcamí, A. Biodiversity and distribution of polar freshwater DNA viruses. Sci. Adv. 2015, 1, e1400127. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-X.; Méheust, R.; Crits-Christoph, A.; McMahon, K.D.; Nelson, T.C.; Slater, G.F.; Warren, L.A.; Banfield, J.F. Large freshwater phages with the potential to augment aerobic methane oxidation. Nat. Microbiol. 2020, 5, 1504–1515. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.A. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 1991, 19, 5442. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New Hhpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [Green Version]
- Laanto, E.; Sundberg, L.-R.; Bamford, J.K.H. Phage specificity of the freshwater fish pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 2011, 77, 7868–7872. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.A.; Heo, J.; Kim, T.-W.; Sang, M.-K.; Song, J.; Kwon, S.-W.; Weon, H.-Y. Xylophilus rhododendri sp. nov., Isolated from Flower of Royal Azalea, Rhododendron schlippenbachii. Curr. Microbiol. 2020, 77, 4160–4166. [Google Scholar] [CrossRef] [PubMed]
- Bowers, L.E.; Weaver, R.H.; Grula, E.A.; Edwards, O.F. Studies on a strain of Caulobacter from water. I. Isolation and identification as Caulobacter vibrioides Henrici and Johnson with emended description. J. Bacteriol. 1954, 68, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.D.; Siddiqi, M.Z.; Liu, Q.; Jo, J.H.; Chun, S.Y.; Choi, G.-M.; Kim, S.Y.; Lee, S.Y.; Im, W.-T. Polaromonas ginsengisoli sp. nov., isolated from ginseng field soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 1436–1441. [Google Scholar] [CrossRef]
- Irgens, R.L.; Gosink, J.J.; Staley, J.T. Polaromonas vacuolate gen. nov., sp. nov., a psychrophilic, marine, gas vacuolate bacterium from Antarctica. Int. J. Syst. Bacteriol. 1996, 46, 822–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, C.O.; Park, W.; Ghiorse, W.C.; Madsen, E.L. Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int. J. Syst. Evol. Microbiol. 2004, 54, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Lokareddy, R.K.; Hou, C.-F.D.; Li, F.; Yang, R.; Cingolani, G. Viral small terminase: A divergent structural framework for a conserved biological function. Viruses 2022, 14, 2215. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Cowan, D.A. Using signature genes as tools to assess environmental viral ecology and diversity. Appl. Environ. Microbiol. 2014, 80, 4470–4480. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, J.; Dreiseikelmann, B.; Rohde, C.; Rohde, M.; Sikorski, J. Isolation and characterization of numerous novel phages targeting diverse strains of the ubiquitous and opportunistic pathogen Achromobacter xylosoxidans. PLoS ONE 2014, 9, e86935. [Google Scholar] [CrossRef]
- Dreiseikelmann, B.; Bunk, B.; Spröer, C.; Rohde, M.; Nimtz, M.; Wittmann, J. Characterization and genome comparisons of three Achromobacter phages of the family Siphoviridae. Arch. Virol. 2017, 162, 2191–2201. [Google Scholar] [CrossRef]
- Skennerton, C.T.; Angly, F.E.; Breitbart, M.; Bragg, L.; He, S.; McMahon, K.D.; Hugenholtz, P.; Tyson, G.W. Phage encoded H-NS: A potential achilles heel in the bacterial defence system. PLoS ONE 2011, 6, e20095. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Plancarte, I.; Cazares, A.; Guarneros, G. Genomic and Transcriptional Mapping of PaMx41, Archetype of a New Lineage of Bacteriophages Infecting Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2016, 82, 6541–6547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.-P.; Tian, F.; Roux, S.; Gazitúa, M.C.; Solonenko, N.E.; Li, Y.-F.; Davis, M.E.; Van Etten, J.L.; Mosley-Thompson, E.; Rich, V.I.; et al. Glacier ice archives nearly 15,000-year-old microbes and phages. Microbiome 2021, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A.; Zaremba-Niedzwiedzka, K.; Martínez-García, M.; McMahon, K.D.; Stepanauskas, R.; Andersson, S.G.; Bertilsson, S. Productivity and salinity structuring of the microplankton revealed by comparative freshwater metagenomics. Environ. Microbiol. 2014, 16, 2682–2698. [Google Scholar] [CrossRef] [Green Version]
- Sepúlveda-Robles, O.; Kameyama, L.; Guarneros, G. High diversity and novel species of Pseudomonas aeruginosa bacteriophages. Appl. Environ. Microbiol. 2012, 78, 4510–4515. [Google Scholar] [CrossRef] [Green Version]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in Aquatic Ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [Green Version]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef] [Green Version]
- Mäntynen, S.; Laanto, E.; Oksanen, H.M.; Poranen, M.M.; Díaz-Muñoz, S.L. Black box of phage-bacterium interactions: Exploring alternative phage infection strategies. Open Biol. 2021, 11, 210188. [Google Scholar] [CrossRef]
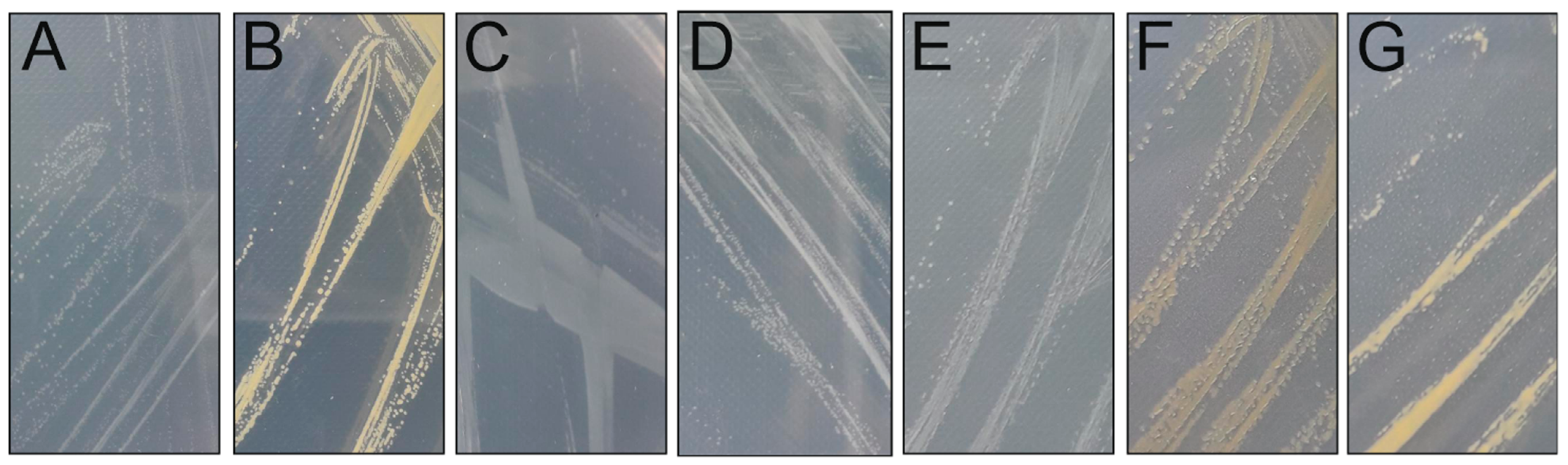

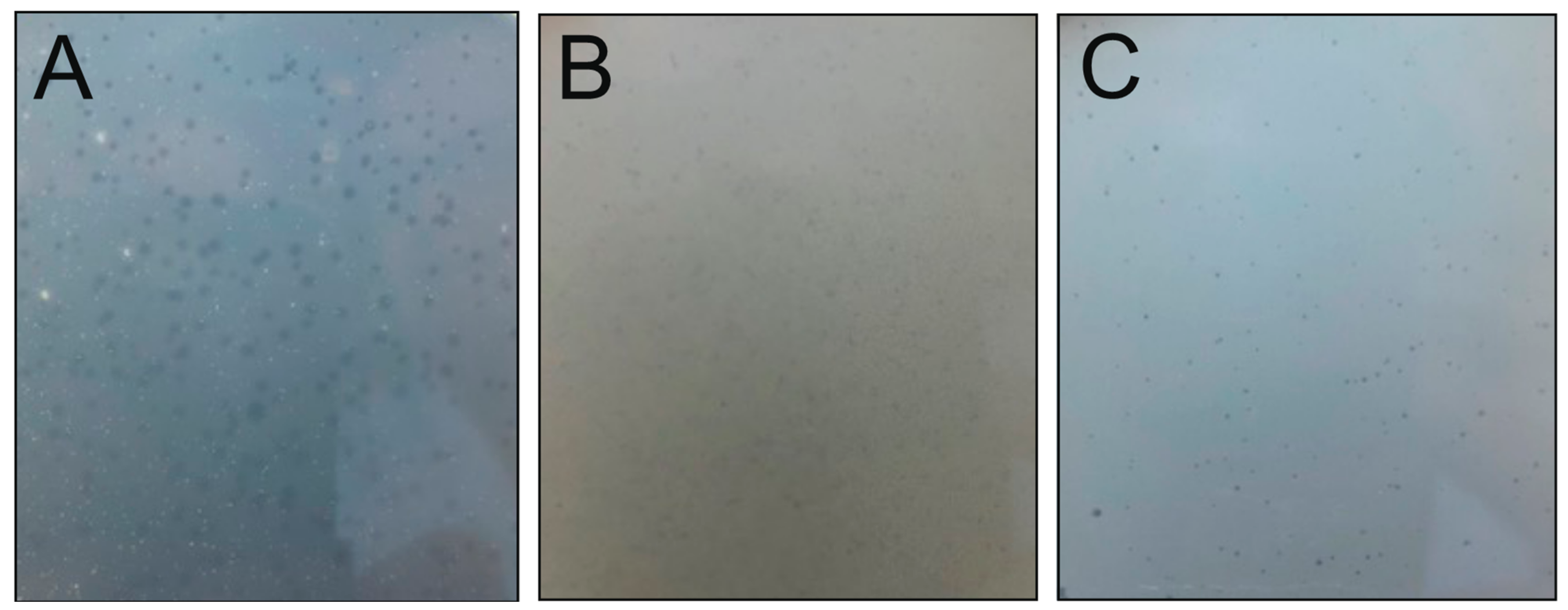
| Strain | Colony Properties | 16S rRNA Partial Sequence Accession ID and Length (bp) | Reference |
|---|---|---|---|
| Xylophilus sp. B14 | Pale, opalescent | OP984743 (1429) | This study |
| Caulobacter sp. B15 | Matte, yellow | OP984744 (1381) | This study |
| Polaromonas sp. B16 | Greenish, pale, matte | OP984745(1424) | This study |
| Pseudomonas sp. B20 | Greenish, pale, matte | OP984746 (478) | This study |
| Herbaspirillum sp. B21 | Pale, opalescent | OP984747 (464) | This study |
| Flavobacterium sp. B28 | Transparent, yellow | FR696328 (853) | [23] |
| Sphingomonas sp. B54 | Matte, yellow | OP984748 (363) | This study |
| Phage 1 | Lumi | Kuura | Tiera |
|---|---|---|---|
| Host bacteria | Xylophilus sp. B14 | Caulobacter sp. B15 | Polaromonas sp. B16 |
| Plaque diameter (mm) 2 | 1–2 | 0.5 | 0.5–1 |
| Titer of virus stock at RT (pfu/mL) 3 | 5.4 × 109 ± 5.0 × 109 | 2.7 × 1010 ± 1.5 × 1010 | 5.0 × 109 ± 1.0 × 109 |
| Virion morphology | siphovirus | podovirus | podovirus |
| Genome length (bp) | 80,496 | 43,205 | 45,327 |
| %GC | 57 | 63 | 48 |
| Genome sequence accession number | OQ067477 | OQ067476 | OQ067478 |
| Reference | This study | This study | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laanto, E.; Oksanen, H.M. Three Phages from a Boreal Lake during Ice Cover Infecting Xylophilus, Caulobacter, and Polaromonas Species. Viruses 2023, 15, 307. https://doi.org/10.3390/v15020307
Laanto E, Oksanen HM. Three Phages from a Boreal Lake during Ice Cover Infecting Xylophilus, Caulobacter, and Polaromonas Species. Viruses. 2023; 15(2):307. https://doi.org/10.3390/v15020307
Chicago/Turabian StyleLaanto, Elina, and Hanna M. Oksanen. 2023. "Three Phages from a Boreal Lake during Ice Cover Infecting Xylophilus, Caulobacter, and Polaromonas Species" Viruses 15, no. 2: 307. https://doi.org/10.3390/v15020307
APA StyleLaanto, E., & Oksanen, H. M. (2023). Three Phages from a Boreal Lake during Ice Cover Infecting Xylophilus, Caulobacter, and Polaromonas Species. Viruses, 15(2), 307. https://doi.org/10.3390/v15020307







