A Glimpse at the Anti-Phage Defenses Landscape in the Foodborne Pathogen Salmonella enterica subsp. enterica serovar Typhimurium
Abstract
:1. Introduction
2. Materials and Methods
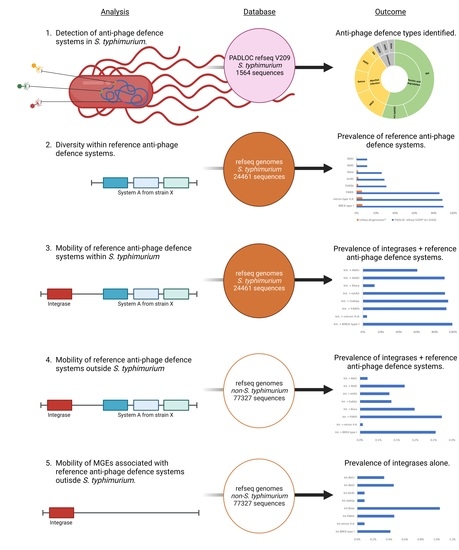
2.1. Detection of Anti-Phage Defense Systems Using the Webserver PADLOC
2.2. Reference Sequences of S. Typhimurium Anti-Phage Defense Systems
2.3. Presence of Reference Anti-phage Defense Systems in S. Typhimurium
2.4. Presence of Reference Anti-phage Defense Systems Co-occurring with Integrases
2.5. Presence of Reference Anti-phage Defense Systems and Their Integrases in Bacterial Species Other than Salmonella Typhimurium
2.6. Presence of Integrases Alone from Reference Anti-phage Defense Systems in Non-Salmonella Typhimurium Bacterial Species
3. Results
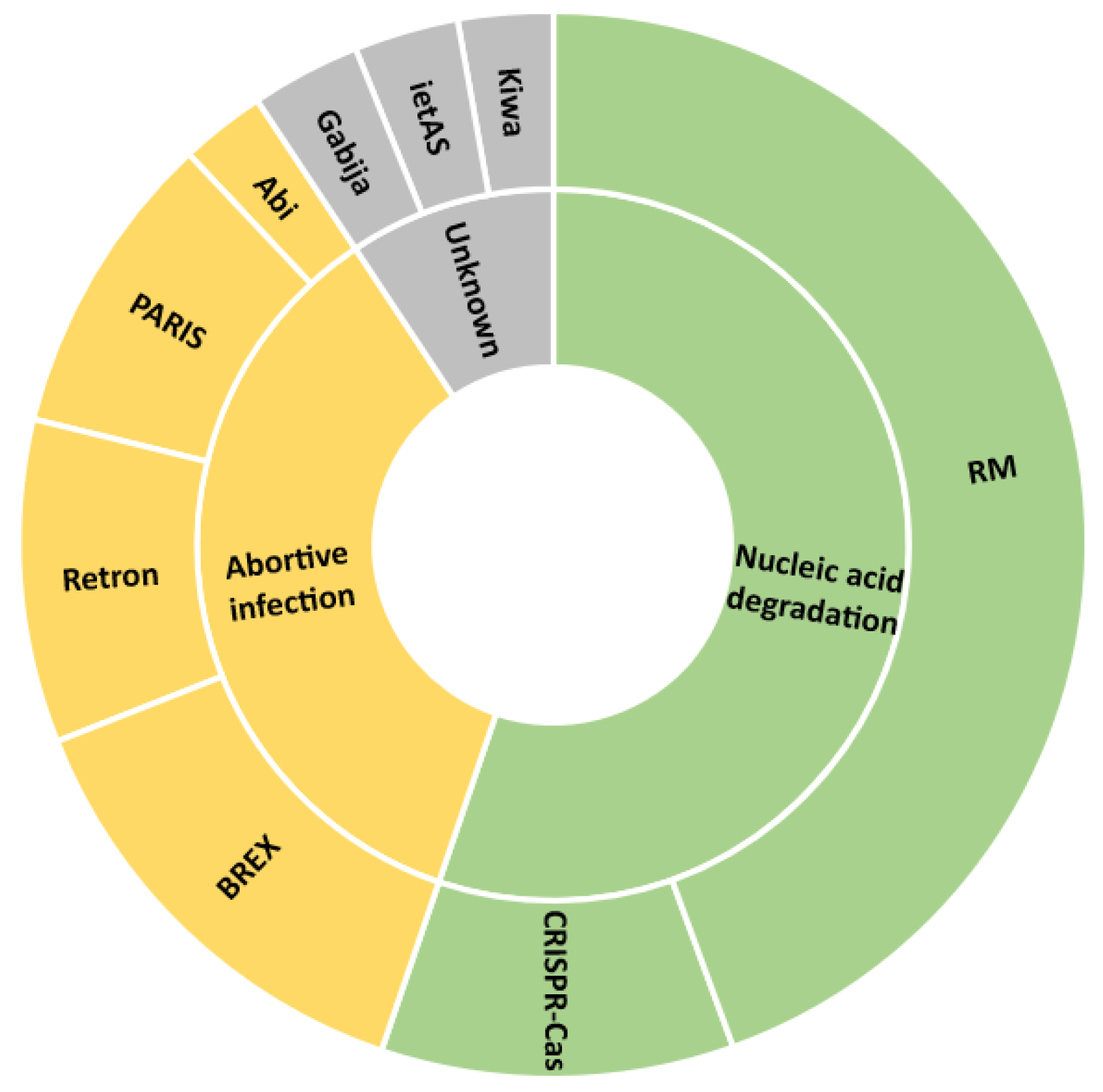
3.1. S. Typhimurium Uses Mainly Nucleic Acid Degradation and Abortive Infection Anti-Phage Defense System Types
3.2. S. Typhimurium Harbors Genetic Diversity within Each Type of Anti-phage Defense System
3.3. S. Typhimurium Anti-Phage Defense Systems Can Be Associated with Diverse Integrases
3.4. S. Typhimurium Integrases Associated with Anti-phage Defense Systems Are found Mainly in Salmonella Enterica Species
3.5. Integrases Alone from Reference Anti-phage Defense Systems Are Found Mainly in Salmonella and in Other g-Proteobacteria Species
4. Discussion
4.1. S. Typhimurium and its Phage Defenses
4.2. S. Typhimurium Carries its Phage Defenses on Mobile Genetic Elements
4.3. Implications of the Mobility of Anti-phage Defense Systems in S. Typhimurium for Using Bacteriophages as Food and Feed Additives
4.4. Are Phage Defense Systems Possibly Involved in Salmonella Epidemiological Success?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wotzka, S.Y.; Nguyen, B.D.; Hardt, W.D. Salmonella Typhimurium Diarrhea Reveals Basic Principles of Enteropathogen Infection and Disease-Promoted DNA Exchange. Cell Host Microbe. 2017, 21, 443–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease “Burden of Illness” Studies. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [Green Version]
- EFSA; ECDC. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Hoffmann, S.; Ahn, J.-W. USDA ERS—Economic Cost of Major Foodborne Illnesses Increased $2 Billion From 2013 to 2018. Available online: https://www.ers.usda.gov/amber-waves/2021/april/economic-cost-of-major-foodborne-illnesses-increased-2-billion-from-2013-to-2018/ (accessed on 1 December 2022).
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The Role of Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Hugas, M.; A Beloeil, P. Controlling Salmonella along the food chain in the European Union—Progress over the last ten years. Eurosurveillance 2014, 19, 20804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wernicki, A.; Nowaczek, A.; Urban-Chmiel, R. Bacteriophage therapy to combat bacterial infections in poultry. Virol. J. 2017, 14, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Au, A.; Lee, H.; Ye, T.; Dave, U.; Rahman, A. Bacteriophages: Combating Antimicrobial Resistance in Food-Borne Bacteria Prevalent in Agriculture. Microorganisms 2022, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef]
- Islam, M.; Zhou, Y.; Liang, L.; Nime, I.; Liu, K.; Yan, T.; Wang, X.; Li, J. Application of a Phage Cocktail for Control of Salmonella in Foods and Reducing Biofilms. Viruses 2019, 11, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Wang, W.; Zhang, Z.; Gu, Y.; Huang, A.; Wang, J.; Hao, H. Phage Products for Fighting Antimicrobial Resistance. Microorganisms 2022, 10, 1324. [Google Scholar] [CrossRef]
- Hagens, S.; De Vegt, B.; Peterson, R. Efficacy of a Commercial Phage Cocktail in Reducing Salmonella Contamination on Poultry Products- Laboratory Data and Industrial Trial Data. Meat Muscle Biol. 2018, 2, 156. [Google Scholar] [CrossRef]
- Bampidis, V.; Azimonti, G.; de Bastos, M.L.; Christensen, H.; Dusemund, B.; Kouba, M.; Durjava, M.F.; López-Alonso, M.; López Puente, S.; Marcon, F.; et al. F/00097 Infecting Salmonella Gallinarum B/00111 (Bafasal®) for All Avian Species (Proteon Pharmaceuticals S.A.). EFSA J. 2021, 19, e06534. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, E.; Stańczyk, M.; Wojtasik, A.; Kowalska, J.; Nowakowska, M.; Łukasiak, M.; Bartnicka, M.; Kazimierczak, J.; Dastych, J. Comprehensive Evaluation of the Safety and Efficacy of BAFASAL® Bacteriophage Preparation for the Reduction of Salmonella in the Food Chain. Viruses 2020, 12, 742. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, V.; Baquero, D.; Hernandez, S.; Farfan, J.; Arias, J.; Arévalo, A.; Donado-Godoy, P.; Vives-Flores, M. Phage cocktail SalmoFREE® reduces Salmonella on a commercial broiler farm. Poult. Sci. 2019, 98, 5054–5063. [Google Scholar] [CrossRef]
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef]
- Rocha, E.P.C.; Bikard, D. Microbial defenses against mobile genetic elements and viruses: Who defends whom from what? PLOS Biol. 2022, 20, e3001514. [Google Scholar] [CrossRef]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef] [Green Version]
- Millman, A.; Melamed, S.; Leavitt, A.; Doron, S.; Bernheim, A.; Hör, J.; Garb, J.; Bechon, N.; Brandis, A.; Lopatina, A.; et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 2022, 30, 1556–1569.e5. [Google Scholar] [CrossRef]
- Gao, L.; Altae-Tran, H.; Böhning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef]
- Hussain, F.A.; Dubert, J.; Elsherbini, J.; Murphy, M.; VanInsberghe, D.; Arevalo, P.; Kauffman, K.; Rodino-Janeiro, B.K.; Gavin, H.; Gomez, A.; et al. Rapid evolutionary turnover of mobile genetic elements drives bacterial resistance to phages. Science 2021, 374, 488–492. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef] [PubMed]
- Pettengill, J.B.; Timme, R.E.; Barrangou, R.; Toro, M.; Allard, M.W.; Strain, E.; Musser, S.M.; Brown, E.W. The evolutionary history and diagnostic utility of the CRISPR-Cas system within Salmonella enterica ssp. enterica. Peerj 2014, 2, e340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roer, L.; Hendriksen, R.S.; Leekitcharoenphon, P.; Lukjancenko, O.; Kaas, R.S.; Hasman, H.; Aarestrup, F.M. Is the Evolution of Salmonella enterica subsp. enterica Linked to Restriction-Modification Systems? Msystems 2016, 1, e00009-16. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res 2008, 36. [Google Scholar] [CrossRef]
- Tal, N.; Sorek, R. SnapShot: Bacterial immunity. Cell 2022, 185, 578. [Google Scholar] [CrossRef]
- Payne, L.J.; Meaden, S.; Mestre, M.R.; Palmer, C.; Toro, N.; Fineran, P.C.; A Jackson, S. PADLOC: A web server for the identification of antiviral defence systems in microbial genomes. Nucleic Acids Res. 2022, 50, W541–W550. [Google Scholar] [CrossRef]
- Payne, L.J.; Todeschini, T.C.; Wu, Y.; Perry, B.J.; Ronson, C.W.; Fineran, P.C.; Nobrega, F.L.; A Jackson, S. Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types. Nucleic Acids Res. 2021, 49, 10868–10878. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Birkholz, N.; A Jackson, S.; Fagerlund, R.D.; Fineran, P.C. A mobile restriction–modification system provides phage defence and resolves an epigenetic conflict with an antagonistic endonuclease. Nucleic Acids Res. 2022, 50, 3348–3361. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; van den Hurk, A.; Aparicio-Maldonado, C.; Krishnakant Kushwaha, S.; King, C.M.; Ou, Y.; Todeschini, T.C.; Clokie, M.R.J.; Millard, A.D.; Gençay, Y.E.; et al. Defense Systems Provide Synergistic Anti-Phage Activity in E. Coli. bioRxiv 2022, 2022–2028. [Google Scholar] [CrossRef]
- Cheng, R.; Huang, F.; Wu, H.; Lu, X.; Yan, Y.; Yu, B.; Wang, X.; Bin Zhu, B. A nucleotide-sensing endonuclease from the Gabija bacterial defense system. Nucleic Acids Res. 2021, 49, 5216–5229. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.H.; Tanji, Y. Bacteriophage-host arm race: An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 2019, 103, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Dai, N.; I Müller, S.; Guan, C.; Parker, M.J.; E Fraser, M.; E Walsh, S.; Sridar, J.; Mulholland, A.; Nayak, K.; et al. Pathways of thymidine hypermodification. Nucleic Acids Res. 2022, 50, 3001–3017. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Dai, N.; Walsh, S.E.; Müller, S.; Fraser, M.E.; Kauffman, K.M.; Guan, C.; Corrêa, I.C., Jr.; Weigele, P.R. Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc. Natl. Acad. Sci. USA 2018, 115, E3116–E3125. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Akusobi, C.; Fang, X.; Salmond, G.P.C. Environmental T4-Family Bacteriophages Evolve to Escape Abortive Infection via Multiple Routes in a Bacterial Host Employing “Altruistic Suicide” through Type III Toxin-Antitoxin Systems. Front. Microbiol. 2017, 8, 1006. [Google Scholar] [CrossRef] [Green Version]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; D’Humières, C.; Cury, J.; Bernheim, A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 2022, 13, 1–10. [Google Scholar] [CrossRef]
- Botelho, J.; Cazares, A.; Schulenburg, H. The ESKAPE Mobilome Contributes to the Spread of Antimicrobial Resistance and CRISPR-Mediated Conflict between Mobile Genetic Elements. Nucleic Acids Res. 2023, 51, 236–252. [Google Scholar] [CrossRef]
- Redondo-Salvo, S.; Fernández-López, R.; Ruiz, R.; Vielva, L.; DE Toro, M.; Rocha, E.P.C.; Garcillán-Barcia, M.P.; De La Cruz, F. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 2020, 11, 3602. [Google Scholar] [CrossRef] [PubMed]
- Cummins, M.L.; Hamidian, M.; Djordjevic, S.P. Salmonella Genomic Island 1 is Broadly Disseminated within Gammaproteobacteriaceae. Microorganisms 2020, 8, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guédon, G.; Libante, V.; Coluzzi, C.; Payot, S.; Leblond-Bourget, N. The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes 2017, 8, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botelho, J.; Schulenburg, H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends Microbiol. 2021, 29, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Delavat, F.; Miyazaki, R.; Carraro, N.; Pradervand, N.; van der Meer, J.R. The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 2017, 41, 512–537. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Bogaj, K.; Bortolaia, V.; Olsen, J.E.; Thomsen, L.E. Antibiotic-Induced, Increased Conjugative Transfer Is Common to Diverse Naturally Occurring ESBL Plasmids in Escherichia coli. Front. Microbiol. 2019, 10, 2119. [Google Scholar] [CrossRef] [PubMed]
- LeGault, K.N.; Hays, S.G.; Angermeyer, A.; McKitterick, A.C.; Johura, F.-T.; Sultana, M.; Ahmed, T.; Alam, M.; Seed, K.D. Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts. Science 2021, 373, eabg2166. [Google Scholar] [CrossRef]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial Retrons Function In Anti-Phage Defense. Cell 2020, 183, 1551–1561.e12. [Google Scholar] [CrossRef]
- Rousset, F.; Depardieu, F.; Miele, S.; Dowding, J.; Laval, A.-L.; Lieberman, E.; Garry, D.; Rocha, E.P.; Bernheim, A.; Bikard, D. Phages and their satellites encode hotspots of antiviral systems. Cell Host Microbe 2022, 30, 740–753.e5. [Google Scholar] [CrossRef]
- Chaudhary, K. BacteRiophage EXclusion (BREX): A Novel Anti-Phage Mechanism in the Arsenal of Bacterial Defense System. J. Cell Physiol. 2018, 233, 771–773. [Google Scholar]
- Johnson, C.M.; Harden, M.M.; Grossman, A.D. Interactions between mobile genetic elements: An anti-phage gene in an integrative and conjugative element protects host cells from predation by a temperate bacteriophage. PLOS Genet. 2022, 18, e1010065. [Google Scholar] [CrossRef] [PubMed]
- Neuert, S.; Nair, S.; Day, M.R.; Doumith, M.; Ashton, P.M.; Mellor, K.C.; Jenkins, C.; Hopkins, K.; Woodford, N.; De Pinna, E.; et al. Prediction of Phenotypic Antimicrobial Resistance Profiles From Whole Genome Sequences of Non-typhoidal Salmonella enterica. Front. Microbiol. 2018, 9, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, M.; Orzechowska, B. Characterisation of Phage Susceptibility Variation in Salmonella enterica Serovar Typhimurium DT104 and DT104b. Microorganisms 2021, 9, 865. [Google Scholar] [CrossRef] [PubMed]
- Kilcher, S.; Loessner, M.J. Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 2019, 27, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Du, J.; Dunne, M.; Kilcher, S.; Loessner, M.J. Engineering therapeutic phages for enhanced antibacterial efficacy. Curr. Opin. Virol. 2022, 52, 182–191. [Google Scholar] [CrossRef]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knøchel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela (Saldinger), S.; Tresse, O. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control, E.F.S.A. Multi-Country Outbreak of Monophasic Salmonella Typhimurium Sequence Type 34 Infections Linked to Chocolate Products, First Update. Available online: https://www.ecdc.europa.eu/en/publications-data/rapid-outbreak-assessment-multi-country-salmonella-outbreak-first-update (accessed on 1 December 2022).
- Le Hello, S.; Hendriksen, R.S.; Doublet, B.; Fisher, I.; Nielsen, E.M.; Whichard, J.M.; Bouchrif, B.; Fashae, K.; Granier, S.A.; Silva, N.J.-D.; et al. International Spread of an Epidemic Population of Salmonella enterica Serotype Kentucky ST198 Resistant to Ciprofloxacin. J. Infect. Dis. 2011, 204, 675–684. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, K.L.; Kirchner, M.; Guerra, B.; Granier, S.A.; Lucarelli, C.; Porrero, M.C.; Jakubczak, A.; Threlfall, E.J.; Mevius, D.J. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: A new pandemic strain? Eurosurveillance 2010, 15, 19580. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef]
| Anti-Phage Defense System | Presence within PADLOC Refseq v209 * (n = 1564) | Accession Number of S. Typhimurium Strain Harboring ref. Defense Systems | Presence of ref. Defense Systems in Refseq Genomes * (NCBI; n = 24,461) |
|---|---|---|---|
| BREX type I | 90% | NZ_JACYBM010000004.1 | 6% |
| retron type II-A | 89% | NZ_JABBKQ010000016.1 | 6% |
| PARIS | 86% | NZ_CP044198.1 | 6% |
| Gabija | 31% | NZ_JABBKQ010000016.1 | 2% |
| ietAS | 29% | NZ_JABBKQ010000016.1 | 2% |
| Kiwa | 26% | NZ_CP050753.1 | 2% |
| AbiD | 11% | NZ_JACXZY010000007.1 | 1% |
| AbiU | 11% | NZ_CP007523.1 | 0.3% |
| Ref. Anti-Phage System Accession Number | Refseq Genomes * | ||
|---|---|---|---|
| Int. + System | MGE Type | Percentage Compared to Positives in Table 1 | |
| Int. + BREX type I | NZ_JACYBM010000004.1 | probable IME | 100% |
| Int. + retron II-A | NZ_JABBKQ010000016.1 | ICE | 5% |
| Int. + PARIS | NZ_CP044198.1 | prophage | 93% |
| Int. + Gabija | NZ_JABBKQ010000016.1 | ICE | 95% |
| Int. + ietAS | NZ_JABBKQ010000016.1 | ICE | 91% |
| Int. + Kiwa | NZ_CP050753.1 | prophage | 13% |
| Int. + AbiD | NZ_JACXZY010000007.1 | transposon | 91% |
| Int. + AbiU | NZ_CP007523.1 | prophage | 61% |
| MGE-int. + Anti-Phage Defense System | Accession Number | BLASTn Count | Presence in Refseq Genomes * (n = 77,327) | Detected Species (% of Hits) |
|---|---|---|---|---|
| Int. + BREX type I | NZ_JACYBM010000004.1 | 159 | 0.2% | Salmonella enterica 100% |
| Int. + retron II-A | NZ_JABBKQ010000016.1 | 6 | 0.0% | Salmonella enterica 100% |
| Int. + PARIS | NZ_CP044198.1 | 172 | 0.2% | Salmonella enterica 99%, Enterobacter 1% |
| Int. + Kiwa | NZ_CP050753.1 | 115 | 0.1% | Salmonella enterica 100% |
| Int. + Gabija | NZ_JABBKQ010000016.1 | 66 | 0.1% | Salmonella enterica 56%, Proteus 33%, Vibrio 5%, other 6% |
| Int. + ietAS | NZ_JABBKQ010000016.1 | 62 | 0.1% | Salmonella enterica 56%, Proteus 34%, Vibrio 5%, other 5% |
| Int. + AbiD | NZ_JACXZY010000007.1 | 94 | 0.1% | Salmonella enterica 78%, Escherichia coli 22% |
| Int. + AbiU | NZ_CP007523.1 | 16 | 0.0% | Salmonella enterica 100% |
| MGE-Int. | Accession Number | BLASTn Count | Presence in Refseq Genomes * (n = 77,327) | Detected Species (% of Hits) |
|---|---|---|---|---|
| Int.BREX type I | NZ_JACYBM010000004.1 | 324 | 0.4% | Other Salmonella 64%, Escherichia 24%, Klebsiella 9%, other 3% |
| Int.retron II-A | NZ_JABBKQ010000016.1 | 70 | 0.1% | Other Salmonella 63%, Proteus 24%, Klebsiella 4%, Shewanella 3%, other 6% |
| Int.PARIS | NZ_CP044198.1 | 361 | 0.5% | Other Salmonella 54%, Klebsiella 16%, Escherichia 15%, Enterobacter 11%, other 4% |
| Int.Gabija | NZ_CP050753.1 | 806 | 1.0% | Other Salmonella 89%, Enterobacter 8%, Citrobacter 3% |
| Int.ietAS | NZ_JABBKQ010000016.1 | 70 | 0.1% | Other Salmonella 63%, Proteus 24%, Klebsiella 4%, Shewanella 3%, other 6% |
| Int.Kiwa | NZ_JABBKQ010000016.1 | 70 | 0.1% | Other Salmonella 63%, Proteus 24%, Klebsiella 4%, Shewanella 3%, other 6% |
| Int.AbiD | NZ_JACXZY010000007.1 | 356 | 0.5% | Escherichia 75%, other Salmonella 21%, Shigella 4% |
| Int.AbiU | NZ_CP007523.1 | 263 | 0.3% | Other Salmonella 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woudstra, C.; Granier, S.A. A Glimpse at the Anti-Phage Defenses Landscape in the Foodborne Pathogen Salmonella enterica subsp. enterica serovar Typhimurium. Viruses 2023, 15, 333. https://doi.org/10.3390/v15020333
Woudstra C, Granier SA. A Glimpse at the Anti-Phage Defenses Landscape in the Foodborne Pathogen Salmonella enterica subsp. enterica serovar Typhimurium. Viruses. 2023; 15(2):333. https://doi.org/10.3390/v15020333
Chicago/Turabian StyleWoudstra, Cedric, and Sophie A. Granier. 2023. "A Glimpse at the Anti-Phage Defenses Landscape in the Foodborne Pathogen Salmonella enterica subsp. enterica serovar Typhimurium" Viruses 15, no. 2: 333. https://doi.org/10.3390/v15020333
APA StyleWoudstra, C., & Granier, S. A. (2023). A Glimpse at the Anti-Phage Defenses Landscape in the Foodborne Pathogen Salmonella enterica subsp. enterica serovar Typhimurium. Viruses, 15(2), 333. https://doi.org/10.3390/v15020333