From Field Tests to Molecular Tools—Evaluating Diagnostic Tests to Improve Rabies Surveillance in Namibia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Rabies Surveillance
2.2. Evaluation of Lateral Flow Devices (LFD) in the Lab and in the Field
2.3. Performance of Commercial RT-PCRs
2.4. Development of a Strain-Specific RT-PCR
2.5. Data Analysis
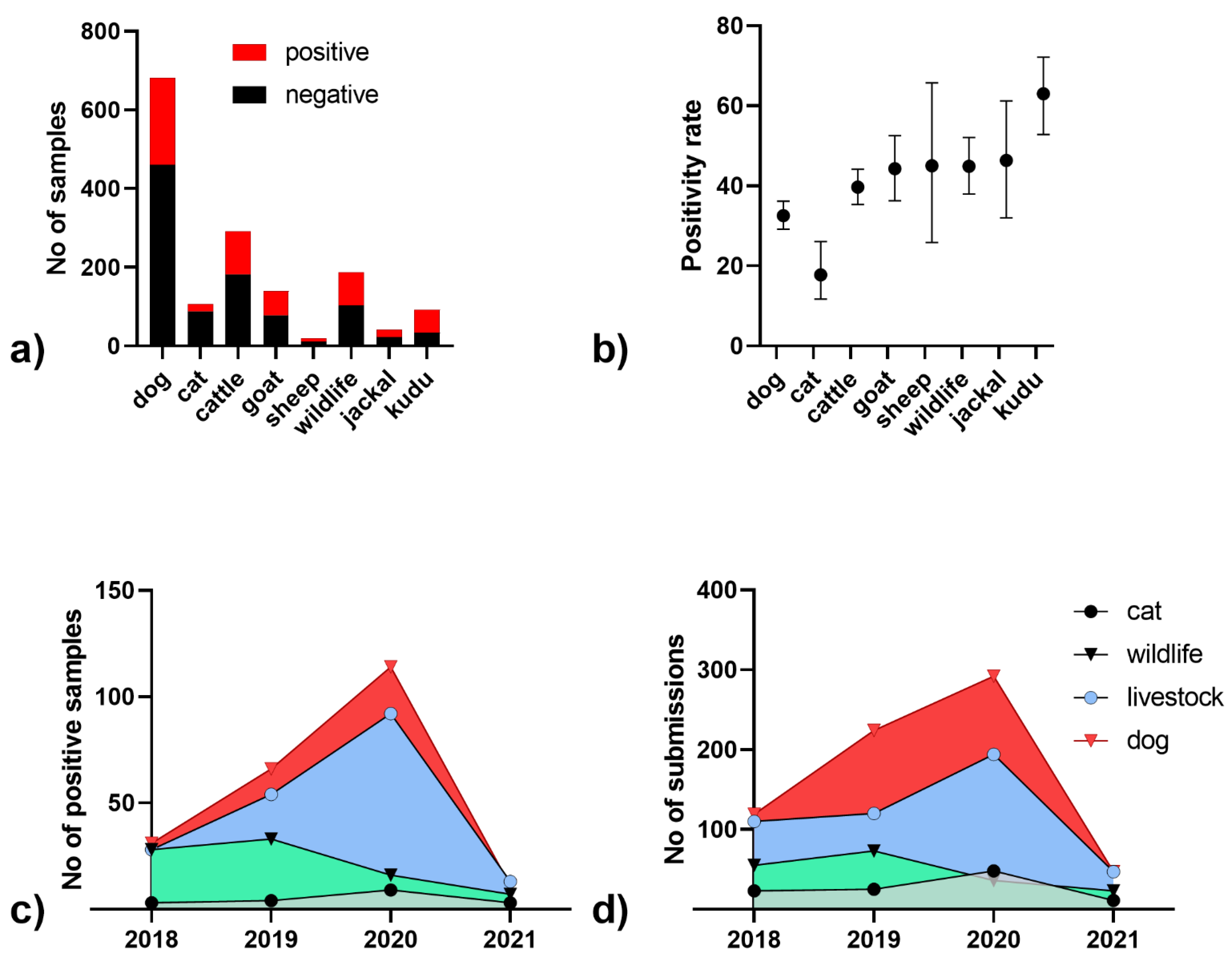
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hikufe, E.H.; Freuling, C.M.; Athingo, R.; Shilongo, A.; Ndevaetela, E.-E.; Helao, M.; Shiindi, M.; Hassel, R.; Bishi, A.; Khaiseb, S.; et al. Ecology and epidemiology of rabies in humans, domestic animals and wildlife in Namibia, 2011-2017. PLoS Neglect. Trop. Dis. 2019, 13, e0007355. [Google Scholar] [CrossRef] [PubMed]
- Hassel, R.H. Incidence of Rabies in Kudu in South West-Africa Namibia. S. Afr. J. Sci. 1982, 78, 418–421. [Google Scholar]
- Barnard, B.J.H.; Hassel, R.H.; Geyer, H.J.; de Koker, W.C. Non-bite transmission of rabies in kudu (Tragelaphus strepsiceros). Onderstepoort J. Vet. Res. 1982, 49, 191–192. [Google Scholar] [PubMed]
- Hassel, R.; Vos, A.; Clausen, P.; Moore, S.; van der Westhuizen, J.; Khaiseb, S.; Kabajani, J.; Pfaff, F.; Höper, D.; Hundt, B.; et al. Experimental screening studies on rabies virus transmission and oral rabies vaccination of the Greater Kudu (Tragelaphus strepsiceros). Sci. Rep. 2018, 8, 16599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, T.P.; Fischer, M.; Khaiseb, S.; Freuling, C.; Höper, D.; Hoffmann, B.; Markotter, W.; Müller, T.; Nel, L.H. Complete genome and molecular epidemiological data infer the maintenance of rabies among kudu (Tragelaphus strepsiceros) in Namibia. PLoS One 2013, 8, e58739. [Google Scholar] [CrossRef]
- Müller, T.; Hassel, R.; Jago, M.; Khaiseb, S.; van der Westhuizen, J.; Vos, A.; Calvelage, S.; Fischer, S.; Marston, D.A.; Fooks, A.R.; et al. Rabies in kudu: Revisited. Adv. Virus Res. 2022, 112, 115–173. [Google Scholar] [CrossRef] [PubMed]
- Athingo, R.; Tenzin, T.; Shilongo, A.; Hikufe, E.; Shoombe, K.K.; Khaiseb, S.; van der Westhuizen, J.; Letshwenyo, M.; Torres, G.; Mettenleiter, T.C.; et al. Fighting Dog-Mediated Rabies in Namibia-Implementation of a Rabies Elimination Program in the Northern Communal Areas. Trop. Med. Infect. Dis. 2020, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- WOAH (Ed.) Terrestrial Animal Health Code; WOAH: Paris, France, 2021; Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/ (accessed on 1 July 2022).
- Fahrion, A.S.; Taylor, L.H.; Torres, G.; Müller, T.; Durr, S.; Knopf, L.; de Balogh, K.; Nel, L.H.; Gordoncillo, M.J.; Abela-Ridder, B. The Road to Dog Rabies Control and Elimination-What Keeps Us from Moving Faster? Front. Public Health 2017, 5, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abela-Ridder, B.; Balogh de, K.; Kessels, J.A.; Dieuzy-Labaye, I.; Torres, G. Global rabies control: The role of international organisations and the Global Strategic Plan to eliminate dog-mediated human rabies. Rev. Sci. Tech. 2018, 37, 741–749. [Google Scholar] [CrossRef] [PubMed]
- WHO. The direct fluorescent antibody test. In Laboratory Techniques in Rabies, 5th ed.; Rupprecht, C.E., Fooks, A.R., Abela-Ridder, B., Eds.; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- OIE. Chapter 2.1.17. Rabies (infection with rabies virus and other lyssaviruses) (NB: Version adopted in May 2018). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2018. [Google Scholar]
- Barrat, J. Simple technique for the collection and shipment of brain specimens for rabies diagnosis. In Laboratory techniques in rabies, 4th; Meslin, F.X., Kaplan, M.M., Koprowski, H., Eds.; World Health Organization: Geneva, Switzerland, 1996; pp. 425–432. [Google Scholar]
- Hoffmann, B.; Freuling, C.M.; Wakeley, P.R.; Rasmussen, T.B.; Leech, S.; Fooks, A.R.; Beer, M.; Müller, T. Improved Safety for Molecular Diagnosis of Classical Rabies Viruses by Use of a TaqMan Real-Time Reverse Transcription-PCR “Double Check” Strategy. J. Clin. Microbiol 2010, 48, 3970–3978. [Google Scholar] [CrossRef] [Green Version]
- Franka, R.; Wallace, R. Rabies diagnosis and surveillance in animals in the era of rabies elimination. Rev. Sci. Tech. 2018, 37, 359–370. [Google Scholar] [CrossRef]
- Nadal, D.; Beeching, S.; Cleaveland, S.; Cronin, K.; Hampson, K.; Steenson, R.; Abela-Ridder, B. Rabies and the pandemic: Lessons for One Health. Trans. R. Soc. Trop. Med. Hyg. 2021, 116, 197–200. [Google Scholar] [CrossRef]
- McElhinney, L.M.; Marston, D.A.; Brookes, S.M.; Fooks, A.R. Effects of carcase decomposition on rabies virus infectivity and detection. J. Virol. Methods 2014, 207C, 110–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WOAH (Ed.) Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2021; WOAH: Paris, France, 2021. [Google Scholar]
- Fooks, A.R.; Johnson, N.; Freuling, C.M.; Wakeley, P.R.; Banyard, A.C.; McElhinney, L.M.; Marston, D.A.; Dastjerdi, A.; Wright, E.; Weiss, R.A.; et al. Emerging technologies for the detection of rabies virus: Challenges and hopes in the 21st century. PLoS Neglect. Trop. Dis. 2009, 3, e530. [Google Scholar] [CrossRef] [Green Version]
- Dacheux, L.; Wacharapluesadee, S.; Hemachudha, T.; Meslin, F.X.; Buchy, P.; Reynes, J.M.; Bourhy, H. More accurate insight into the incidence of human rabies in developing countries through validated laboratory techniques. PLoS Neglect. Trop. Dis. 2010, 4, e765. [Google Scholar] [CrossRef] [Green Version]
- Eggerbauer, E.; de Benedictis, P.; Hoffmann, B.; Mettenleiter, T.C.; Schlottau, K.; Ngoepe, E.C.; Sabeta, C.T.; Freuling, C.M.; Müller, T. Evaluation of Six Commercially Available Rapid Immunochromatographic Tests for the Diagnosis of Rabies in Brain Material. PLoS Neglect. Trop. Dis. 2016, 10, e0004776. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Fahrion, A.; Finke, S.; Eyngor, M.; Novak, S.; Yakobson, B.; Ngoepe, E.; Phahladira, B.; Sabeta, C.; de Benedictis, P.; et al. Further Evidence of Inadequate Quality in Lateral Flow Devices Commercially Offered for the Diagnosis of Rabies. Trop. Med. Infect. Dis. 2020, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Layan, M.; Dellicour, S.; Baele, G.; Cauchemez, S.; Bourhy, H. Mathematical modelling and phylodynamics for the study of dog rabies dynamics and control: A scoping review. PLoS Neglect. Trop. Dis. 2021, 15, e0009449. [Google Scholar] [CrossRef]
- Brunker, K.; Marston, D.A.; Horton, D.L.; Cleaveland, S.; Fooks, A.R.; Kazwala, R.; Ngeleja, C.; Lembo, T.; Sambo, M.; Mtema, Z.J.; et al. Elucidating the phylodynamics of endemic rabies virus in eastern Africa using whole-genome sequencing. Virus Evol. 2015, 1, vev011. [Google Scholar] [CrossRef] [Green Version]
- Brunker, K.; Jaswant, G.; Thumbi, S.M.; Lushasi, K.; Lugelo, A.; Czupryna, A.M.; Ade, F.; Wambura, G.; Chuchu, V.; Steenson, R.; et al. Rapid in-country sequencing of whole virus genomes to inform rabies elimination programmes. Wellcome Open Res 2020, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Gigante, C.M.; Yale, G.; Condori, R.E.; Costa, N.C.; van Long, N.; Minh, P.Q.; Chuong, V.D.; Tho, N.D.; Thanh, N.T.; Thin, N.X.; et al. Portable Rabies Virus Sequencing in Canine Rabies Endemic Countries Using the Oxford Nanopore MinION. Viruses 2020, 12, 1255. [Google Scholar] [CrossRef] [PubMed]
- de Pace, V.; Bruzzone, B.; Orsi, A.; Ricucci, V.; Domnich, A.; Guarona, G.; Randazzo, N.; Stefanelli, F.; Battolla, E.; Dusi, P.A.; et al. Comparative Analysis of Five Multiplex RT-PCR Assays in the Screening of SARS-CoV-2 Variants. Microorganisms 2022, 10, 306. [Google Scholar] [CrossRef] [PubMed]
- Mauti, S.; Léchenne, M.; Naïssengar, S.; Traoré, A.; Kallo, V.; Kouakou, C.; Couacy-Hymann, E.; Gourlaouen, M.; Mbilo, C.; Pyana, P.P.; et al. Field Postmortem Rabies Rapid Immunochromatographic Diagnostic Test for Resource-Limited Settings with Further Molecular Applications. J. Vis. Exp. 2020, e60008. [Google Scholar] [CrossRef]
- Yale, G.; Gibson, A.D.; Mani, R.S.; P K, H.; Costa, N.C.; Corfmat, J.; Otter, I.; Otter, N.; Handel, I.G.; Bronsvoort, B.M.; et al. Evaluation of an Immunochromatographic Assay as a Canine Rabies Surveillance Tool in Goa, India. Viruses 2019, 11, 649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenzin, T.; Lhamo, K.; Rai, P.B.; Tshering, D.; Jamtsho, P.; Namgyal, J.; Wangdi, T.; Letho, S.; Rai, T.; Jamtsho, S.; et al. Evaluation of a rapid immunochromatographic test kit to the gold standard fluorescent antibody test for diagnosis of rabies in animals in Bhutan. BMC Vet. Res. 2020, 16, 183. [Google Scholar] [CrossRef]
- Klein, A.; Eggerbauer, E.; Potratz, M.; Zaeck, L.M.; Calvelage, S.; Finke, S.; Müller, T.; Freuling, C.M. Comparative pathogenesis of different phylogroup I bat lyssaviruses in a standardized mouse model. PLoS Neglect. Trop. Dis. 2022, 16, e0009845. [Google Scholar] [CrossRef]


| Assay | Name | Role | Length (nt) | Sequence | Position * | PCR or In Vitro RNA Fragment (bp or nt) |
|---|---|---|---|---|---|---|
| R13 | JW12 | primer | 19 | ATGTAACACCYCTACAATG | 55–73 | 110 |
| N165–146 | primer | 20 | GCAGGGTAYTTRTACTCATA | 165–146 | ||
| LysGT1-FAM | probe | 29 |
6-FAM-
ACAAGATTGTATTCAAAGTCAATAATCAG-TAMRA | 81–109 | ||
| R14 | RV-N_F | primer | 23 | GATCCTGATGAYGTATGTTCCTA | 266–288 | 87 |
| RV-N_R | primer | 19 | RGATTCCGTAGCTRGTCCA | 353–335 | ||
| RabGT1-B-FAM: | probe | 25 | 6-FAM- CAGCAATGCAGTTYTTTGAGGGGAC-TAMRA | 297–321 | ||
| Dog_F1 | Frag_1_F | primer | 20 | GGAGCTGAATAACACGGTGC | 1438–1457 | 85 |
| Frag_1_R | primer | 20 | AACCATCCCAGACATGAGCA | 1508–1489 | ||
| Frag_1_Probe_FAM | probe | 22 | 6-FAM-TGATCGTGCATATCCATCATGA-TAMRA | 1455–1476 |
| (a) | FLI | Total | (b) | RT-qPCR (FLI) | Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| POS | NEG | POS | NEG | ||||||
| CVL | POS | 108 | 5 | 113 | FAT(CVL) | POS | 108 | 5 | 113 |
| NEG | 2 | 111 | 113 | NEG | 12 | 101 | 113 | ||
| total | 110 | 116 | 226 | 120 | 106 | 226 | |||
| Lab-ID | Species | FAT | FLI (R13/14) | Ingenetix | Genesig | PCRMAX | Norgen Biotek | Techne | Liferiver | KogeneBiotech |
|---|---|---|---|---|---|---|---|---|---|---|
| 46864 | jackal | pos | 21.87 | 21.01 | 23.53 | 26.09 | 28.75 | 29.41 | 39.21 | N/A |
| 46869 | jackal | pos | 16.36 | 13.58 | 17.22 | 19.36 | 22.11 | 23.76 | 34.47 | N/A |
| 47142 | eland | pos | 20.01 | 26.24 | 28.33 | 35.30 | 35.12 | 33.81 | 34.25 | N/A |
| 47177 | kudu | pos | 20.91 | 19.18 | 31.01 | 23.04 | 36.15 | 34.67 | N/A | N/A |
| 47198 | kudu | pos | 19.75 | 17.61 | 29.03 | 27.61 | N/A | 34.42 | 34.23 | N/A |
| 47314 | dog | pos | 30.12 | 31.01 | 34.50 | N/A | 38.48 | 24.87 | N/A | N/A |
| 47319 | dog | pos | 15.19 | 15.24 | 18.42 | 20.21 | 23.10 | N/A | 31.07 | N/A |
| 47325 | dog | pos | 18.35 | 20.49 | 25.11 | 26.91 | 31.65 | 26.05 | 37.80 | N/A |
| 47416 | dog | pos | 17.91 | 26.35 | 26.51 | 31.24 | 23.66 | 31.95 | 39.39 | N/A |
| 47536 | goat | pos | 17.25 | 25.46 | 29.47 | 36.18 | 29.31 | N/A | N/A | N/A |
| PC * | 19.75 | 19.70 | 15.75 | 15.43 | 24.23 | 14.60 | 23.77 | 24.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freuling, C.M.; van der Westhuizen, J.; Khaiseb, S.; Tenzin, T.; Müller, T. From Field Tests to Molecular Tools—Evaluating Diagnostic Tests to Improve Rabies Surveillance in Namibia. Viruses 2023, 15, 371. https://doi.org/10.3390/v15020371
Freuling CM, van der Westhuizen J, Khaiseb S, Tenzin T, Müller T. From Field Tests to Molecular Tools—Evaluating Diagnostic Tests to Improve Rabies Surveillance in Namibia. Viruses. 2023; 15(2):371. https://doi.org/10.3390/v15020371
Chicago/Turabian StyleFreuling, Conrad M., Jolandie van der Westhuizen, Siegfried Khaiseb, Tenzin Tenzin, and Thomas Müller. 2023. "From Field Tests to Molecular Tools—Evaluating Diagnostic Tests to Improve Rabies Surveillance in Namibia" Viruses 15, no. 2: 371. https://doi.org/10.3390/v15020371





