Influence of Climatic Variables on Incidence of Whitefly-Transmitted Begomovirus in Soybean and Bean Crops in North-Western Argentina
Abstract
:1. Introduction
2. Materials and Methods
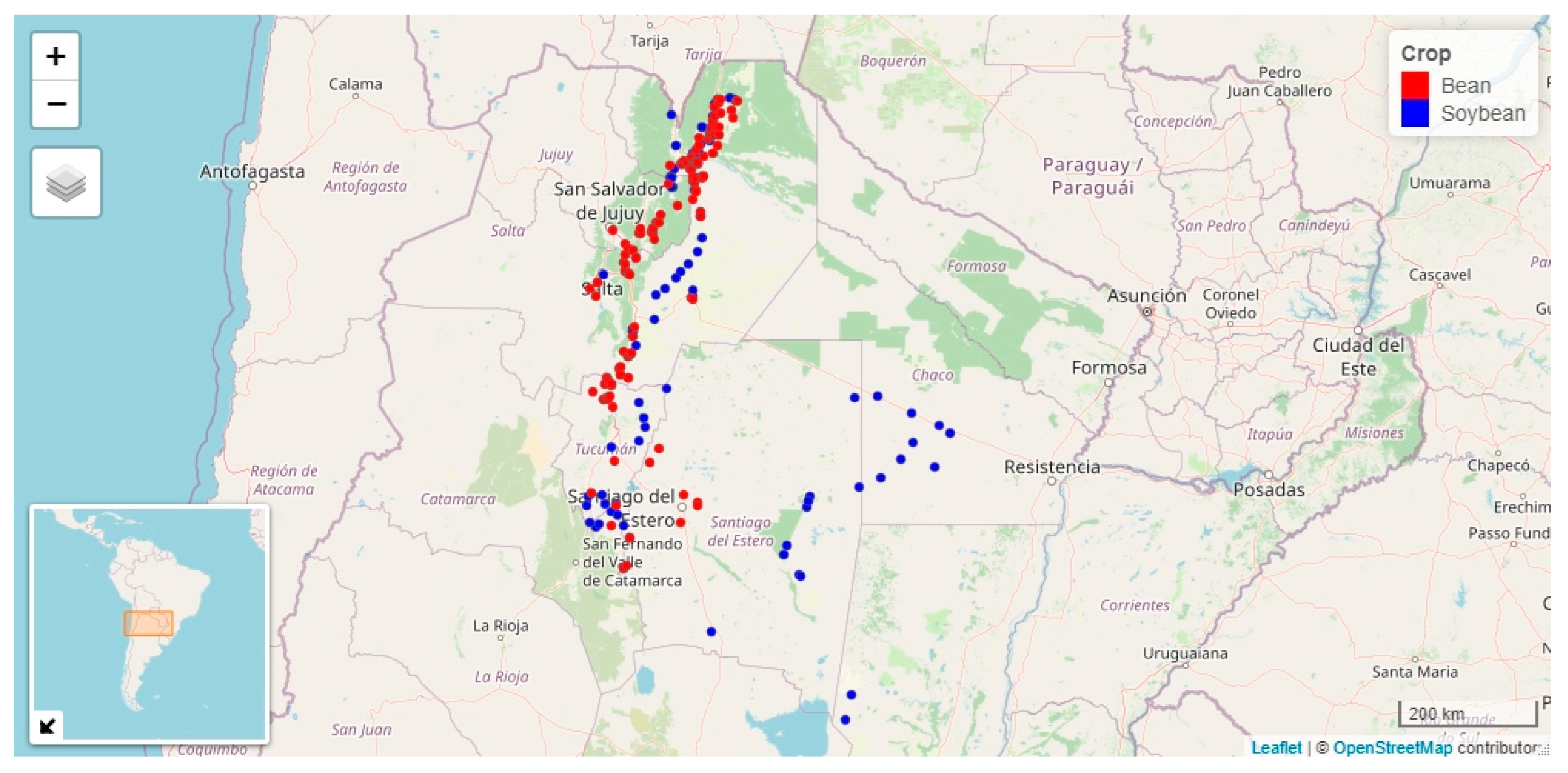
2.1. Plant Sampling and Analysis
2.2. Obtaining Biometeorological Variables
2.2.1. Dataset of Begomovirus in Bean Crop
2.2.2. Dataset of Begomovirus in Soybean Crop
2.3. Statistical Analyses
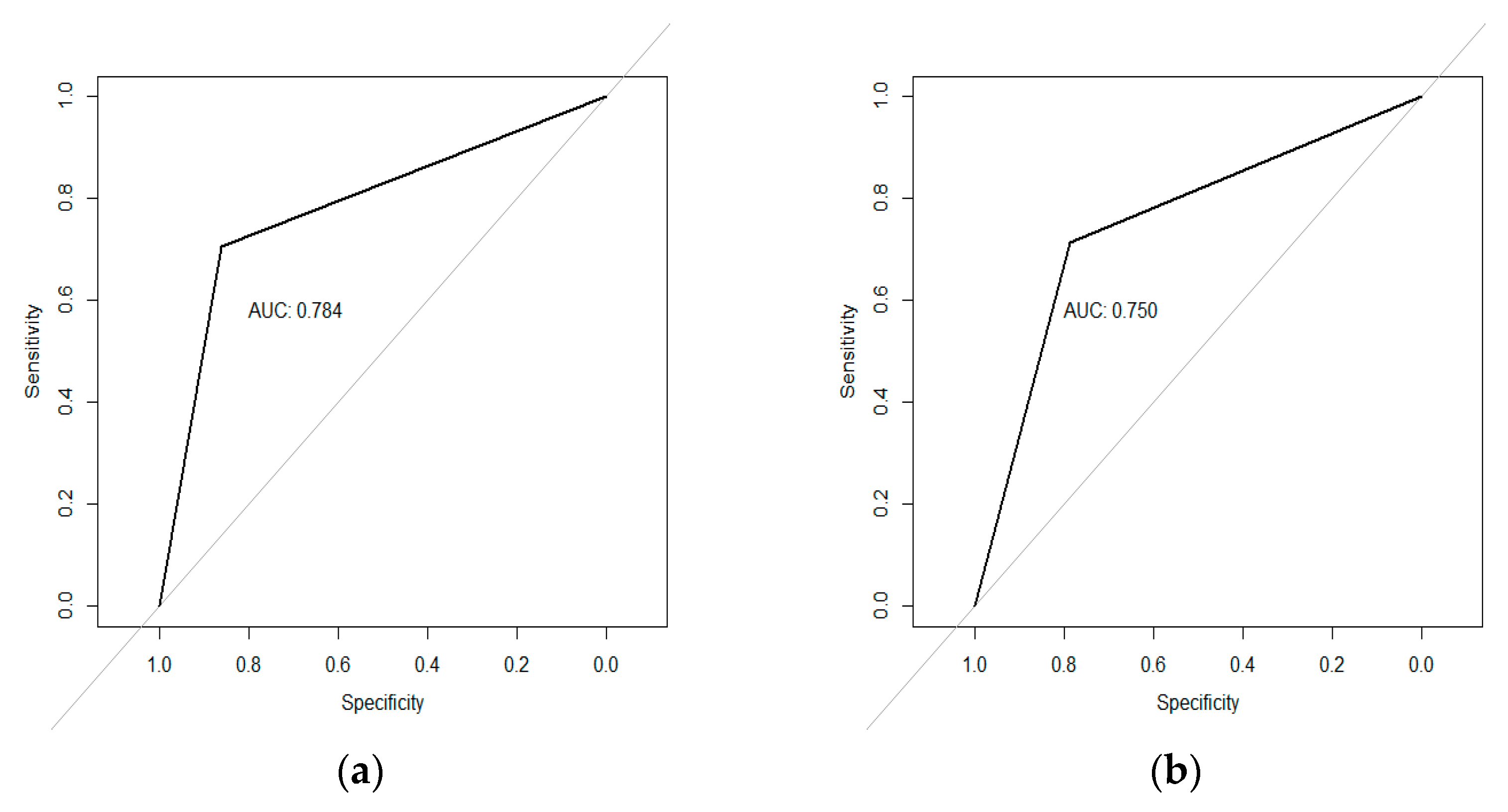
2.4. Model Assesement
2.5. Validation of Predictors
3. Results
3.1. Begomovirus Incidence and Putative Biometeorological Variables
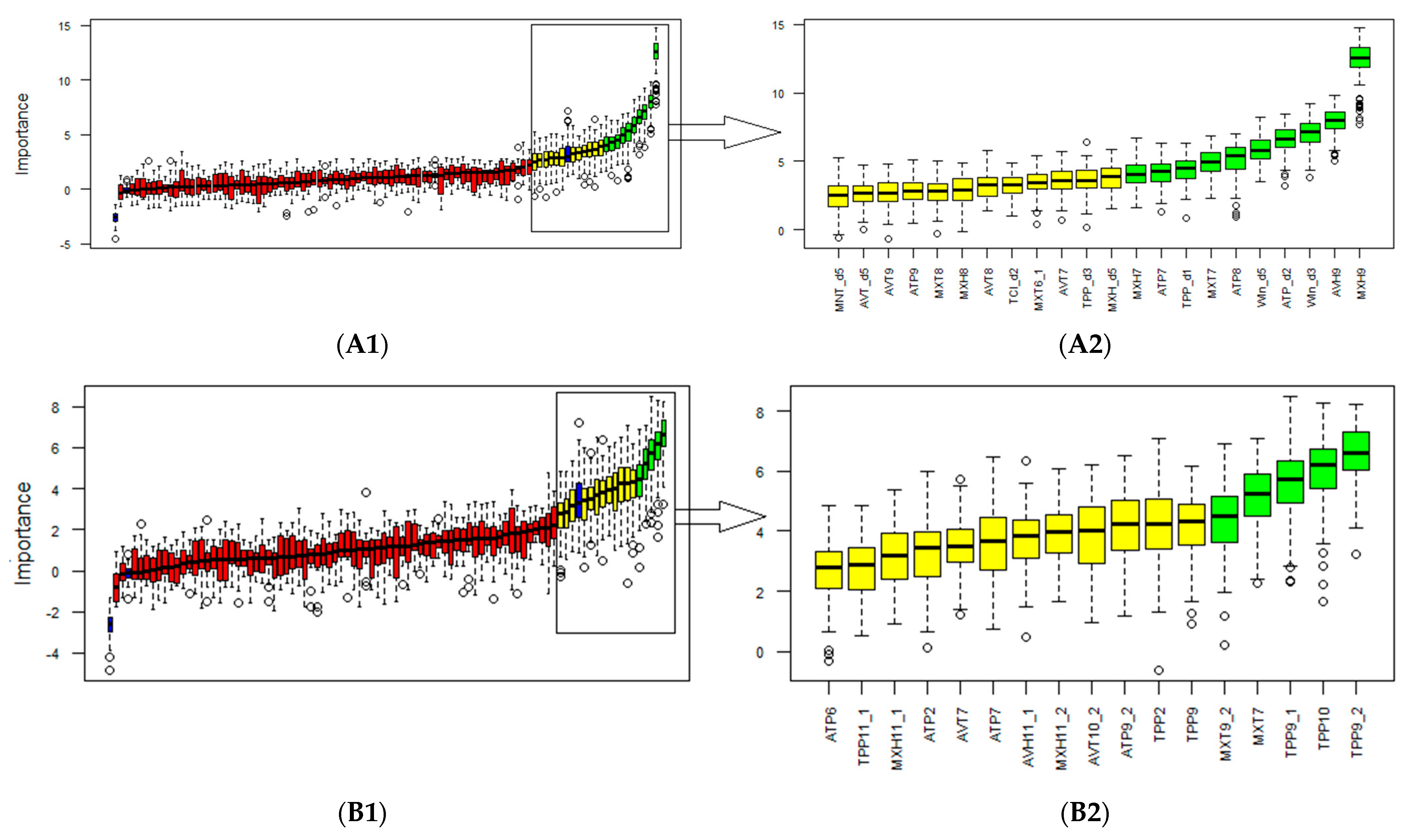
3.2. Selection of Biometeorological Variables
3.3. Regression Models Fitted to Predict Begomovirus Incidence
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Varma, A.; Malathi, V.G. Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 2003, 142, 145–164. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Marchant, W.G.; Gautam, S.; Dutta, B.; Srinivasan, R. Whitefly-Mediated Transmission and Subsequent Acquisition of Highly Similar and Naturally Occurring Tomato Yellow Leaf Curl Virus Variants. Phytopathology® 2022, 112, 720–728. [Google Scholar] [CrossRef]
- Macedo, M.A.; Costa, T.M.; Barbosa, J.C.; Pereira, J.L.; Michereff-Filho, M.; Gilbertson, R.L.; Inoue-Nagata, A.K.; Filho, A.B. Temporal and spatial dynamics of begomovirus disease in tomatoes in central Brazil. Plant Pathol. 2017, 66, 529–538. [Google Scholar] [CrossRef]
- Fernandes, F.R.; Cruz, A.R.R.; Faria, J.C.; Zerbini, F.M.; Aragão, F.J.L. Three distinct begomoviruses associated with soybean in central Brazil. Arch. Virol. 2009, 154, 1567–1570. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Lima, M.F.; Gilbertson, R.L. A review of geminivirus diseases in vegetables and other crops in Brazil: Current status and approaches for management. Hortic. Bras. 2016, 34, 8–18. [Google Scholar] [CrossRef]
- Varela, G.; Ávalos, V.; Reyna, P.; Laguna, I.G.; Pardina, P.R. Identification, molecular characterization and relative incidence of begomoviruses infecting bean crops in northwestern Argentina: An update. Australas. Plant Pathol. 2018, 47, 343–350. [Google Scholar] [CrossRef]
- Morales, F.J. History and Current Distribution of Begomoviruses in Latin America. Adv. Virus Res. 2006, 67, 127–162. [Google Scholar] [CrossRef]
- Gautam, S.; Gadhave, K.R.; Buck, J.W.; Dutta, B.; Coolong, T.; Adkins, S.; Srinivasan, R. Virus-virus interactions in a plant host and in a hemipteran vector: Implications for vector fitness and virus epidemics. Virus Res. 2020, 286, 198069. [Google Scholar] [CrossRef]
- De Barro, P.; Ahmed, M.Z. Genetic networking of the Bemisia tabaci cryptic species complex reveals pattern of biological invasions. PLoS ONE 2011, 6, e25579. [Google Scholar] [CrossRef]
- Shadmany, M.; Boykin, L.M.; Muhamad, R.; Omar, D. Genetic diversity of Bemisia tabaci (Hemiptera: Aleyrodidae) species complex across Malaysia. J. Econ. Entomol. 2019, 112, 75–84. [Google Scholar] [CrossRef]
- Xu, J.; De Barro, P.J.; Liu, S.S. Reproductive incompatibility among genetic groups of Bemisia tabaci supports the proposition that the whitefly is a cryptic species complex. Bull. Entomol. Res. 2010, 100, 359–366. [Google Scholar] [CrossRef]
- Alemandri, V.; Rodriguez Pardina, P.; Argüello Caro, E.; Mattio, M.F.; Dumón, A.; Rodriguez, S.M.; Truol, G.; Izaurralde, J.; García Medina, S. Incidence of begomoviruses and climatic characterisation of Bemisia tabaci-geminivirus complex in soybean and bean in Argentina. Agriscientia 2012, 29, 31–39. [Google Scholar] [CrossRef]
- Alemandri, V.; Vaghi Medina, C.G.; DumÓn, A.D.; Argüello Caro, E.B.; Mattio, M.F.; Garcia Medina, S.; López Lambertini, P.M.; Truol, G. Three members of the Bemisia tabaci (Hemiptera: Aleyrodidae) cryptic species complex occur sympatrically in Argentine horticultural crops. J. Econ. Entomol. 2015, 108, 405–413. [Google Scholar] [CrossRef]
- Krause-Sakate, R.; Watanabe, L.F.M.; Gorayeb, E.S.; da Silva, F.B.; Alvarez, D.D.L.; Bello, V.H.; Nogueira, A.M.; de Marchi, B.R.; Vicentin, E.; Ribeiro-Junior, M.R.; et al. Population dynamics of whiteflies and associated viruses in South America: Research progress and perspectives. Insects 2020, 11, 847. [Google Scholar] [CrossRef]
- Kalyebi, A.; MacFadyen, S.; Parry, H.; Tay, W.T.; De Barro, P.; Colvin, J. African cassava whitefly, Bemisia tabaci, cassava colonization preferences and control implications. PLoS ONE 2018, 13, e0204862. [Google Scholar] [CrossRef]
- Guo, C.L.; Zhu, Y.Z.; Zhang, Y.J.; Keller, M.A.; Liu, T.X.; Chu, D. Invasion Biology and Management of Sweetpotato Whitefly (Hemiptera: Aleyrodidae) in China. J. Integr. Pest Manag. 2021, 12, 2–3. [Google Scholar] [CrossRef]
- Viscarret, M.M.; Torres-Jerez, I.; Agostini de Manero, E.; López, S.N.; Botto, E.E.; Brown, J.K. Mitochondrial DNA Evidence for a Distinct New World Group of Bemisia tabaci (Gennadius) (Hemiptera:Aleyrodidae) Indigenous to Argentina and Bolivia, and Presence of the Old World B Biotype in Argentina. Ann. Entomol. Soc. Am. 2003, 96, 65–72. [Google Scholar] [CrossRef]
- Alemandri, V.; De Barro, P.; Bejerman, N.; Caro, E.B.A.; Dumón, A.D.; Mattio, M.F.; Rodriguez, S.M.; Truol, G. Species Within the Bemisia tabaci (Hemiptera: Aleyrodidae) Complex in Soybean and Bean Crops in Argentina. J. Econ. Entomol. 2012, 105, 48–53. [Google Scholar] [CrossRef]
- Gautam, S.; Crossley, M.S.; Dutta, B.; Coolong, T.; Simmons, A.M.; da Silva, A.; Snyder, W.E.; Srinivasan, R. Low Genetic Variability in Bemisia tabaci MEAM1 Populations within Farmscapes of Georgia, USA. Insects 2020, 11, 834. [Google Scholar] [CrossRef] [PubMed]
- Kriticos, D.J.; Darnell, R.E.; Yonow, T.; Ota, N.; Sutherst, R.W.; Parry, H.R.; Mugerwa, H.; Maruthi, M.N.; Seal, S.E.; Colvin, J.; et al. Improving climate suitability for Bemisia tabaci in East Africa is correlated with increased prevalence of whiteflies and cassava diseases. Sci. Rep. 2020, 10, 22049. [Google Scholar] [CrossRef] [PubMed]
- Morales-Soto, A.; Lamz-Piedra, A. Genetic improvement methods in the cultivation of common beans (Phaseolus vulgaris L.) against the Bean Yellow Golden Mosaic Virus (BGYMV). Cultiv. Trop. 2020, 41, e10. [Google Scholar]
- FAO, F. Food and Agriculture Organization of the United Nations. Rome. 2019. Available online: https://www.fao.org/faostat/en/#home (accessed on 21 September 2022).
- Storti, L. Informes de Cadenas de Valor. Oleaginosas: Soja—Septiembre. 2019. Available online: https://www.argentina.gob.ar/sites/default/files/sspmicro_cadenas_de_valor_soja.pdf (accessed on 19 September 2022).
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Rodríguez-Pardina, P.E.; Hanada, K.; Laguna, I.G.; Zerbini, F.M.; Ducasse, D.A. Molecular characterisation and relative incidence of bean- and soybean-infecting begomoviruses in northwestern Argentina. Ann. Appl. Biol. 2011, 158, 69–78. [Google Scholar] [CrossRef]
- Czernecki, B.; Głogowski, A.; Nowosad, J. Climate: An R package to access free in-situ meteorological and hydrological datasets for environmental assessment. Sustainability 2020, 12, 394. [Google Scholar] [CrossRef]
- Vega, A.; Córdoba, M.; Castro-Franco, M.; Balzarini, M. Protocol for automating error removal from yield maps. Precis. Agric. 2019, 20, 1030–1044. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R.; Kursa, M.M.B. Wrapper Algorithm for All Relevant Feature Selection. Available online: https://gitlab.com/mbq/Boruta/. (accessed on 21 September 2022).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.r-project.org/ (accessed on 21 September 2022).
- Agresti, A.; Kateri, M. Some Remarks on Latent Variable Models in Categorical Data Analysis. Commun. Stat.-Theory Methods 2014, 43, 801–814. [Google Scholar] [CrossRef]
- Kuhn, M. The Caret Package; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: https://cran.r-project.org/web/packages/caret/index.html (accessed on 21 September 2022).
- Carmona, M.; Moschini, R.; Cazenave, G.; Sautua, F. Relación entre la precipitación registrada en estados reproductivos de la soja y la severidad de Septoria glycines y Cercospora kikuchii. Trop. Plant Pathol. 2010, 35, 71–78. Available online: www.sbfito.com.br (accessed on 19 August 2021). [CrossRef]
- Yin, J.; Mutiso, F.; Tian, L. Joint hypothesis testing of the area under the receiver operating characteristic curve and the Youden index. Pharm. Stat. 2021, 20, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression; John Wiley & Sons: New York, NY, USA, 2000. [Google Scholar]
- Evans, J.S.; Murphy, M.A. Package ‘Rfutilities’; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Vizgarra, O.N.; Espeche, C.; Mamani, S. Cultivos de Verano Para El Norte Argentino-Poroto (Phaseolus vulgaris L.): Manejo del Cultivo y Recomendaciones. Av. Agroind. 2012, 3, 29–32. [Google Scholar]
- Vizgarra, O.N.; Espeche, C.; Mamaní Gonzáles, S.; Velazquez, D.; Ploper, L.D. Consideraciones generales de la campana de poroto 2012 y resultados de los ensayos evaluados en el Noroeste Argentino. Av. Agroind. 2012, 33, 29–34. [Google Scholar]
- Vizgarra, O.N.; Ploper, L.D.; Ghio, A.C.; Pérez, A. TUC 510: Nueva variedad de poroto negro (Phaseolus vulgaris L.) con alta tolerancia a las virosis presentes en el noroeste argentino. Rev. Ind. Agrícola Tucumán 2006, 83, 39–47. [Google Scholar]
- Moschini, R.C.; Fortugno, C. Predicting wheat head blight incidence using models based on meteorological factors in Pergamino, Argentina. Eur. J. Plant Pathol. 1996, 102, 211–218. [Google Scholar] [CrossRef]
- Pozzi, E.A.; Bruno, C.; Luciani, C.E.; Celli, M.G.; Conci, V.C.; Perotto, M.C. Relative incidence of cucurbit viruses and relationship with bio-meteorological variables. Australas. Plant Pathol. 2020, 49, 167–174. [Google Scholar] [CrossRef]
- Heinemann, A.B.; Ramirez-Villegas, J.; Souza, T.L.P.O.; Didonet, A.D.; di Stefano, J.G.; Boote, K.J.; Jarvis, A. Drought impact on rainfed common bean production areas in Brazil. Agric. For. Meteorol. 2016, 225, 57–74. [Google Scholar] [CrossRef]
- Field, C.B.; Barros, V.R.; Dokken, D.J.; Mach, K.J.; Mastrandrea, M.D.; Bilir, T.E.; Chatterjee, M.; Ebi, K.L.; Estrada, Y.O.; Genova, R.C.; et al. Available online: https://www.ipcc.ch/site/assets/uploads/2018/02/WGIIAR5-PartA_FINAL.pdf (accessed on 21 September 2022).
- Molina, R.M.O. Eficacia de los Agrotextiles Como Método de Protección de Cultivos; Universidad Almería: Almería, Spain, 2017. [Google Scholar]
- Picó, B.; Díez, M.J.; Nuez, F. Viral diseases causing the greatest economics losses to the tomato crop II. The Tomato yellow leaf curly virus—A review. Sci. Hortic. 1996, 67, 151–196. [Google Scholar] [CrossRef]
- Ghosh, A.; Rao, G.P.; Baranwal, V.K. Manual on Transmission of Plant Viruses and Phytoplasmas by Insect Vectors; Indian Agricultural Research Institute: New Delhi, India, 2019. [Google Scholar]
- Morales, F.J.; Jones, P.G. The ecology and epidemiology of whitefly-transmitted viruses in Latin America. Virus Res. 2004, 100, 57–65. [Google Scholar] [CrossRef]
- March, G.; Balzarini, M.; Ornaghi, J.; Beviacqua, J. Modelo Predictivo de la Intensidad de la Enfermedad “Mal de Río Cuarto”. Enferm. Plantas 1995, 79, 1051–1053. Available online: https://agris.fao.org/agris-search/search.do?recordID=US9563432 (accessed on 3 August 2022).
| Estimate | Std. Error | z Value | Pr(>|z|) | |||
|---|---|---|---|---|---|---|
| Bean | Intercept | 9.0749 | 10.0292 | 0.905 | 0.365545 | |
| MXT6_1 | −0.3599 | 0.1233 | −2.920 | 0.003506 | ** | |
| MXT7 | 2.1496 | 0.7224 | 2.976 | 0.002925 | ** | |
| AVT7 | −2.3241 | 0.6961 | −3.338 | 0.000842 | *** | |
| MXT8 | −2.1989 | 0.5513 | −3.989 | 6.64 × 10−5 | *** | |
| AVT8 | 3.6146 | 0.8607 | 4.200 | 2.67 × 10−5 | *** | |
| AVH9 | −0.4208 | 0.0998 | −4.213 | 2.52 × 10−5 | *** | |
| TPP_d1 | −0.0152 | 0.0071 | −2.132 | 0.033041 | * | |
| Soybean | Intercept | 10.050 | 4.918 | 2.044 | 0.040994 | * |
| AVT9_1 | 0.303 | 0.123 | 2.456 | 0.014043 | * | |
| TPP9_2 | −0.160 | 0.047 | −3.376 | 0.000734 | *** | |
| AVT10_2 | −0.622 | 0.231 | −2.691 | 0.007125 | ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyna, P.; Suarez, F.; Balzarini, M.; Rodriguez Pardina, P. Influence of Climatic Variables on Incidence of Whitefly-Transmitted Begomovirus in Soybean and Bean Crops in North-Western Argentina. Viruses 2023, 15, 462. https://doi.org/10.3390/v15020462
Reyna P, Suarez F, Balzarini M, Rodriguez Pardina P. Influence of Climatic Variables on Incidence of Whitefly-Transmitted Begomovirus in Soybean and Bean Crops in North-Western Argentina. Viruses. 2023; 15(2):462. https://doi.org/10.3390/v15020462
Chicago/Turabian StyleReyna, Pablo, Franco Suarez, Mónica Balzarini, and Patricia Rodriguez Pardina. 2023. "Influence of Climatic Variables on Incidence of Whitefly-Transmitted Begomovirus in Soybean and Bean Crops in North-Western Argentina" Viruses 15, no. 2: 462. https://doi.org/10.3390/v15020462
APA StyleReyna, P., Suarez, F., Balzarini, M., & Rodriguez Pardina, P. (2023). Influence of Climatic Variables on Incidence of Whitefly-Transmitted Begomovirus in Soybean and Bean Crops in North-Western Argentina. Viruses, 15(2), 462. https://doi.org/10.3390/v15020462







