Deubiquitinating Enzyme Inhibitors Block Chikungunya Virus Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Reagents and Antibodies
2.3. Inhibitor Treatment, Cell Viability Evaluation, and CHIKV Infection
2.4. Infected Cell Detection Using Flow Cytometry
2.5. Viral Protein Detection Using Western Blotting
2.6. Viral RNA Quantification Using RT-qPCR
2.7. Viral Titration Using Plaque Assay
2.8. Statistical Analysis
3. Results
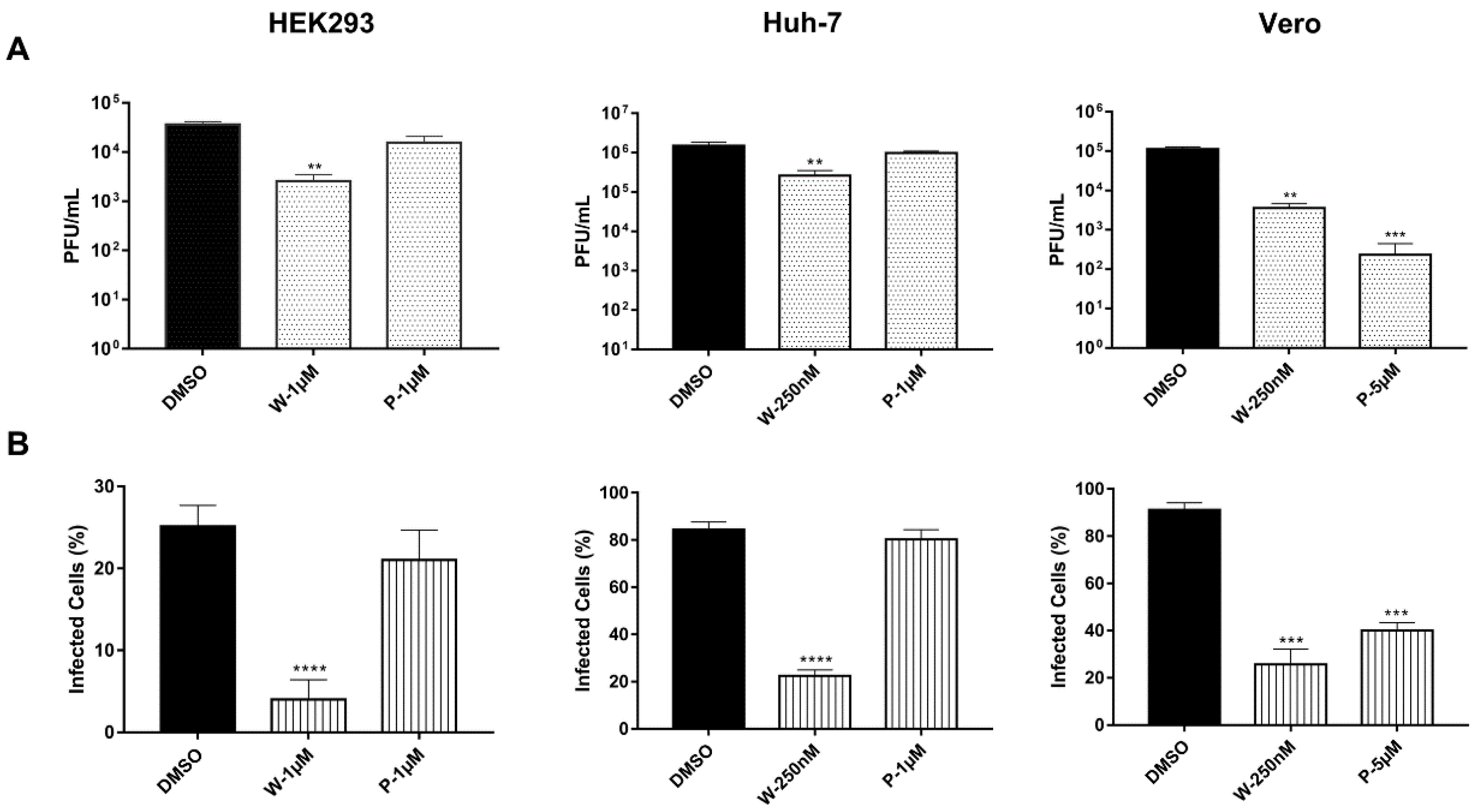
3.1. Chikungunya Virus Requires the Activity of DUBs to Achieve a Productive Infection
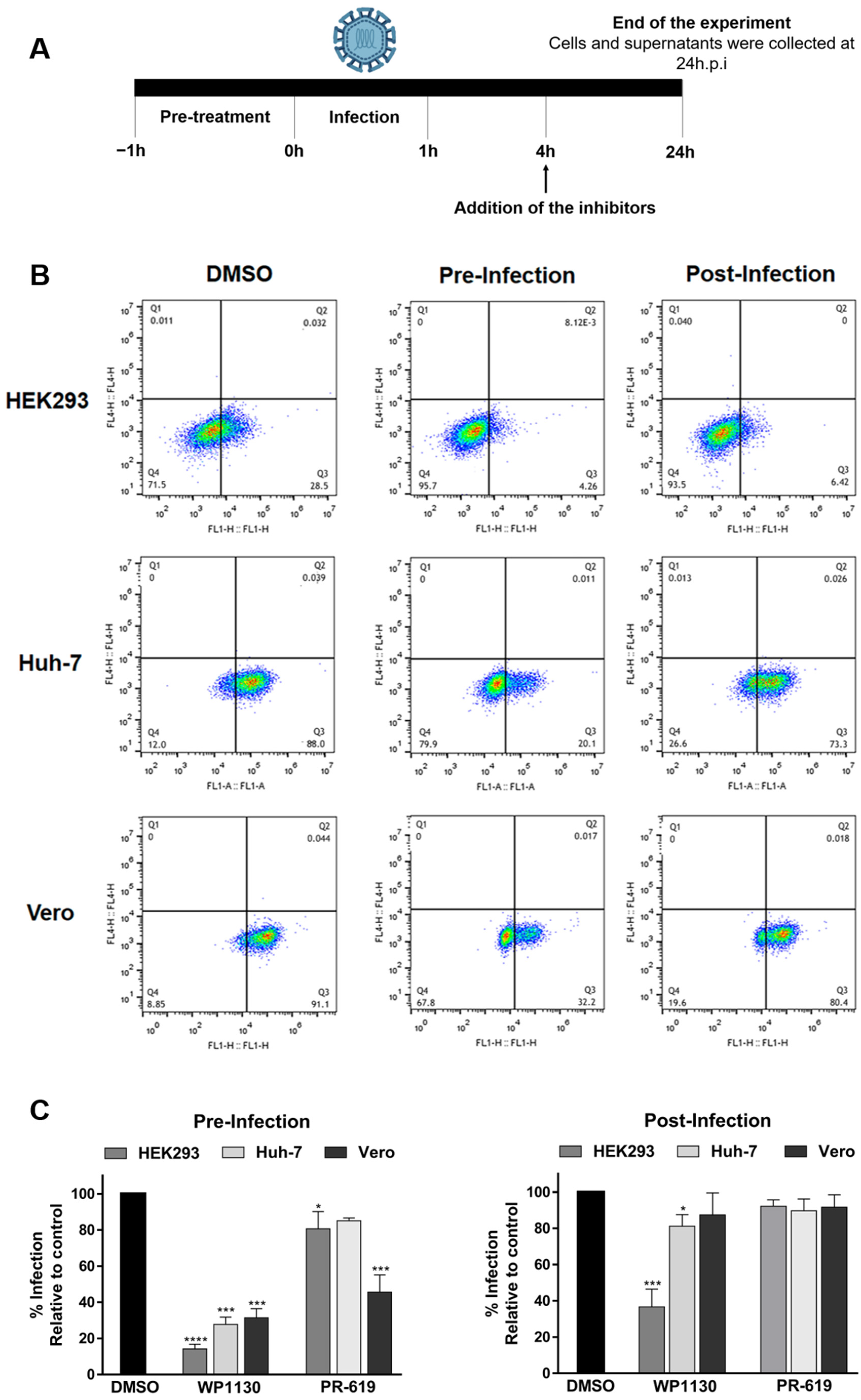
3.2. Inhibition of DUBs Affects Key Events in Viral Replication
3.3. Deubiquitinating Enzymes Are Not Regulated during CHIKV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Li, Y.; Reverter, D. Molecular mechanisms of dubs regulation in signaling and disease. Int. J. Mol. Sci. 2021, 22, 986. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biocheml. Scis. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Valerdi, K.M.; Hage, A.; van Tol, S.; Rajsbaum, R.; Giraldo, M.I. The Role of the Host Ubiquitin System in Promoting Replication of Emergent Viruses. Viruses 2021, 13, 369. [Google Scholar] [CrossRef]
- Gu, H.; Fada, F.J. Specificity in ubiquitination triggered by virus infection. Int. J. Mol. Sci. 2020, 21, 4088. [Google Scholar] [CrossRef]
- Kumar, R.; Mehta, D.; Mishra, N.; Nayak, D.; Sunil, S. Role of host-mediated post-translational modifications (PTMS) in RNA virus pathogenesis. Int. J. Mol. Sci. 2021, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.M.; Lee, B.H.; Nicola, A.V. Viral entry and the ubiquitin-proteasome system. Cell. Microbiol. 2021, 23, e13276. [Google Scholar] [CrossRef]
- Dhanwani, R.; Khan, M.; Lomash, V.; Rao, P.V.L.; Ly, H.; Parida, M. Characterization of chikungunya virus induced host response in a mouse model of viral myositis. PLoS ONE 2014, 9, 1–12. [Google Scholar] [CrossRef]
- Thio, C.P.L.; Yusof, R.; Abdul-Rahman, P.S.A.; Karsani, S.A. Differential Proteome Analysis of Chikungunya Virus Infection on Host Cells. PLoS ONE 2013, 8, e61444. [Google Scholar] [CrossRef]
- Treffers, E.E.; Tas, A.; Scholte, F.E.; Van, M.N.; Heemskerk, M.T.; de Ru, A.H.; Snijder, E.J.; van Hemert, M.J.; van Veelen, P.A. Temporal SILAC-based quantitative proteomics identifies host factors involved in chikungunya virus replication. Proteomics 2015, 15, 2267–2280. [Google Scholar] [CrossRef]
- Karpe, Y.A.; Pingale, K.D.; Kanade, G.D. Activities of proteasome and m-calpain are essential for Chikungunya virus replication. Virus Genes 2016, 52, 716–721. [Google Scholar] [CrossRef]
- Kaur, P.; Lello, L.S.; Utt, A.; Dutta, S.K.; Merits, A.; Chu, J.J.H. Bortezomib inhibits chikungunya virus replication by interfering with viral protein synthesis. PLoS Negl. Trop. Dis. 2020, 14, 1–25. [Google Scholar] [CrossRef]
- Economopoulou, A.; Dominguez, M.; Helynck, B.; Sissoko, D.; Wichmann, O.; Quenel, P.; Germonneau, P.; Quatresous, I. Atypical Chikungunya virus infections: Clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Réunion. Epidemiol. Infect. 2009, 137, 534–541. [Google Scholar] [CrossRef]
- Soumahoro, M.K.; Gérardin, P.; Boelle, P.Y.; Perrau, J.; Fianu, A.; Pouchot, J.; Malvy, D.; Flahault, A.; Favier, F.; Hanslik, T. Impact of Chikungunya virus infection on health status and quality of life: A retrospective cohort study. PLoS ONE 2009, 4, 1–6. [Google Scholar] [CrossRef]
- Cunha, M.S.; Costa, P.A.G.; Correa, I.A.; de Souza, M.R.M.; Calil, P.T.; da Silva, G.P.D.; Costa, S.M.; Fonseca, V.W.P.; da Costa, L.J. Chikungunya Virus: An Emergent Arbovirus to the South American Continent and a Continuous Threat to the World. Front. Microbiol. 2020, 11, 1297. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.A. Weekly Report|PAHO/WHO. In: Pan American Health Organization/World Health Organization [Internet]. 17 January 2019. Available online: https://www3.paho.org/data/index.php/en/mnu-topics/chikv-en/550-chikvweekly-en.html (accessed on 24 November 2022).
- Calvo, E.P.; Archila, E.D.; López, L.; Castellanos, J.E. Chikungunya virus rediscovering. Biomedica 2021, 41, 1–49. [Google Scholar] [CrossRef]
- Archila, E.D.; López, L.S.; Castellanos, J.E.; Calvo, E.P. Molecular and biological characterization of an Asian-American isolate of Chikungunya virus. PLoS ONE 2022, 17, e0266450. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Altun, M.; Kramer, H.B.; Willems, L.I.; McDermott, J.L.; Leach, C.A.; Goldenberg, S.J.; Kumar, K.G.S.; Konietzny, R.; Fischer, R.; Kogan, E.; et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011, 18, 1401–1412. [Google Scholar] [CrossRef]
- Kapuria, V.; Peterson, L.F.; Fang, D.; Bornmann, W.; Talpaz, M.; Donato, N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010, 70, 9265–9276. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Zhang, W.; Gabler, S.; Chipman, P.R.; Strauss, E.G.; Strauss, J.H.; Baker, T.S.; Kuhn, R.J.; Rossmann, M.G. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 2006, 14, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Feibelman, K.M.; Fuller, B.P.; Li, L.; LaBarbera, D.V.; Geiss, B.J. Identification of small molecule inhibitors of the Chikungunya virus nsP1 RNA capping enzyme. Antivir. Res. 2018, 154, 124–131. [Google Scholar] [CrossRef]
- Feng, W.; Sun, X.; Shi, N.; Zhang, M.; Guan, Z.; Duan, M. Influenza a virus NS1 protein induced A20 contributes to viral replication by suppressing interferon-induced antiviral response. Biochem. Biophys. Res. Commun. 2017, 482, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Karim, R.; Tummers, B.; Meyers, C.; Biryukov, J.L.; Alam, S.; Backendorf, C.; Jha, V.; Offringa, R.; van Ommen, G.-J.B.; Melief, C.J.M.; et al. Human Papillomavirus (HPV) Upregulates the Cellular Deubiquitinase UCHL1 to Suppress the Keratinocyte’s Innate Immune Response. PLoS Pathog. 2013, 9, e1003384. [Google Scholar] [CrossRef]
- Yokota, S.; Okabayashi, T.; Yokosawa, N.; Fujii, N. Measles virus P protein suppresses Toll-like receptor signal through up-regulation of ubiquitin-modifying enzyme A20. FASEB J. 2008, 22, 74–83. [Google Scholar] [CrossRef]
- Nair, S.R.; Abraham, R.; Sundaram, S.; Sreekumar, E. Interferon regulated gene (IRG) expression-signature in a mouse model of chikungunya virus neurovirulence. J. Neurovirol. 2017, 23, 886–902. [Google Scholar] [CrossRef] [PubMed]
- Laplante, G.; Zhang, W. Targeting the ubiquitin-proteasome system for cancer therapeutics by small-molecule inhibitors. Cancers 2021, 13, 3079. [Google Scholar] [CrossRef] [PubMed]
- Nag, D.K.; Finley, D. A small-molecule inhibitor of deubiquitinating enzyme USP14 inhibits Dengue virus replication. Virus Res. 2012, 165, 103–106. [Google Scholar] [CrossRef]
- Perry, J.W.; Ahmed, M.; Chang, K.O.; Donato, N.J.; Showalter, H.D.; Wobus, C.E. Antiviral activity of a small molecule deubiquitinase inhibitor occurs via induction of the unfolded protein response. PLoS Pathog. 2012, 8, 43. [Google Scholar] [CrossRef]
- Setz, C.; Friedrich, M.; Rauch, P.; Fraedrich, K.; Matthaei, A.; Traxdorf, M.; Schubert, U. Inhibitors of deubiquitinating enzymes block HIV-1 replication and augment the presentation of gag-derived MHC-I epitopes. Viruses 2017, 9, 222. [Google Scholar] [CrossRef]
- Große, M.; Setz, C.; Rauch, P.; Auth, J.; Morokutti-Kurz, M.; Temchura, V.; Schubert, U. Inhibitors of Deubiquitinating Enzymes Interfere with the SARS-CoV-2 Papain-like Protease and Block Virus Replication In Vitro. Viruses 2022, 14, 1404. [Google Scholar] [CrossRef]
- Ali, A.; Raja, R.; Farooqui, S.R.; Ahmad, S.; Banerjea, A.C. USP7 deubiquitinase controls HIV-1 production by stabilizing Tat protein. Biochem. J. 2017, 474, 1653–1668. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López, L.S.; Calvo, E.P.; Castellanos, J.E. Deubiquitinating Enzyme Inhibitors Block Chikungunya Virus Replication. Viruses 2023, 15, 481. https://doi.org/10.3390/v15020481
López LS, Calvo EP, Castellanos JE. Deubiquitinating Enzyme Inhibitors Block Chikungunya Virus Replication. Viruses. 2023; 15(2):481. https://doi.org/10.3390/v15020481
Chicago/Turabian StyleLópez, Lady S., Eliana P. Calvo, and Jaime E. Castellanos. 2023. "Deubiquitinating Enzyme Inhibitors Block Chikungunya Virus Replication" Viruses 15, no. 2: 481. https://doi.org/10.3390/v15020481




