Altered microRNA Transcriptome in Cultured Human Airway Cells upon Infection with SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions
2.2. Protein Extractions and Western Blot
2.3. RNA Isolation
2.4. Illumina Nextseq Sequencing of Cells Infected with SARS-CoV-2 Viruses
2.5. Go and KEGG Annotation/Enrichment
2.6. RT-qPCR
2.7. Plasmid Constructs
2.8. Cell Transfection and Dual-Luciferase Assay
2.9. Statistical Analysis of qPCR Data
3. Results
3.1. SARS-CoV-2 Replication in Calu-3 Cells and Preliminary Analysis of the Molecular Mechanisms Underlying the Disease
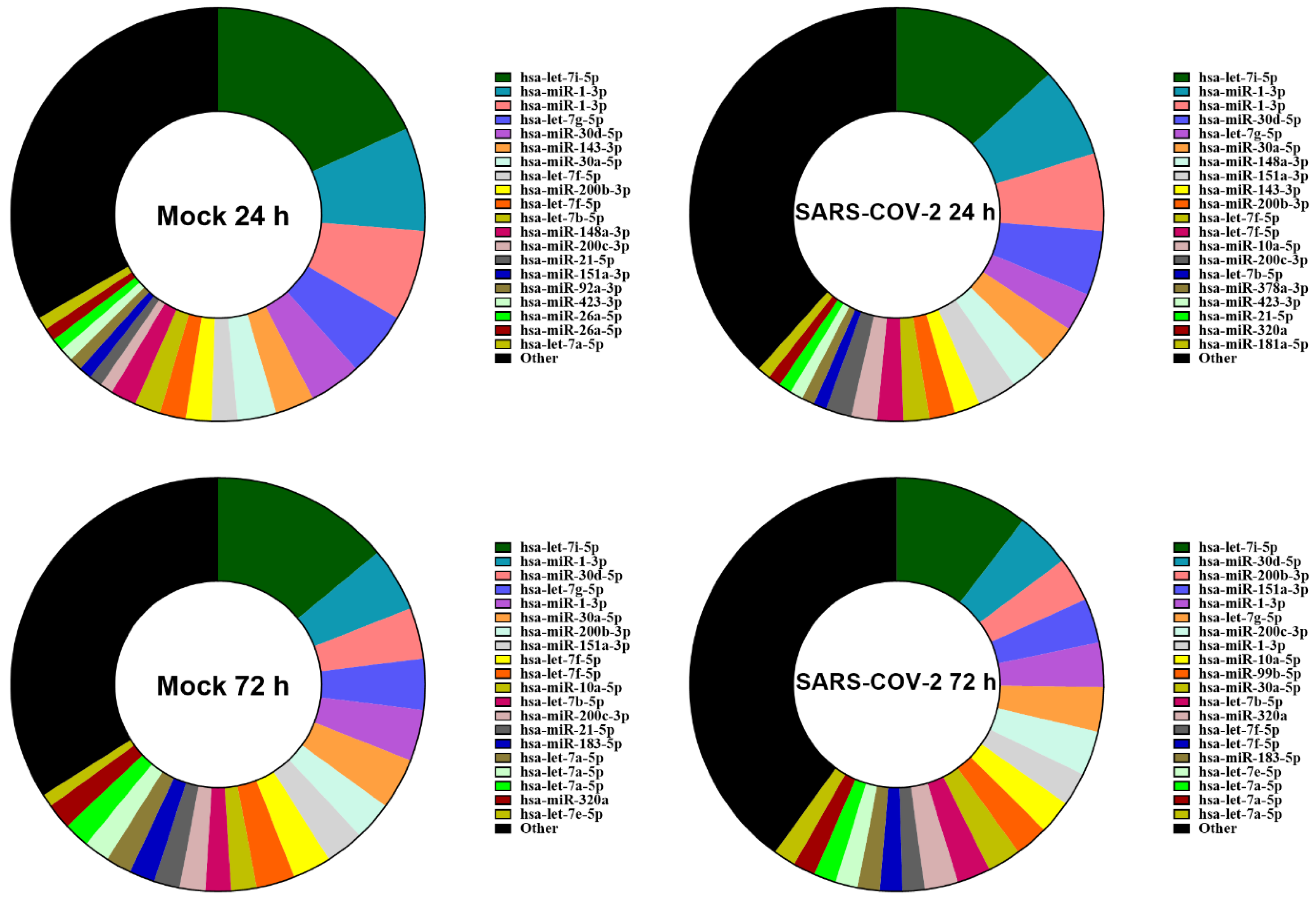
3.2. SARS-CoV-2 Infection Does Not Alter the Relative Abundance of miRNAs
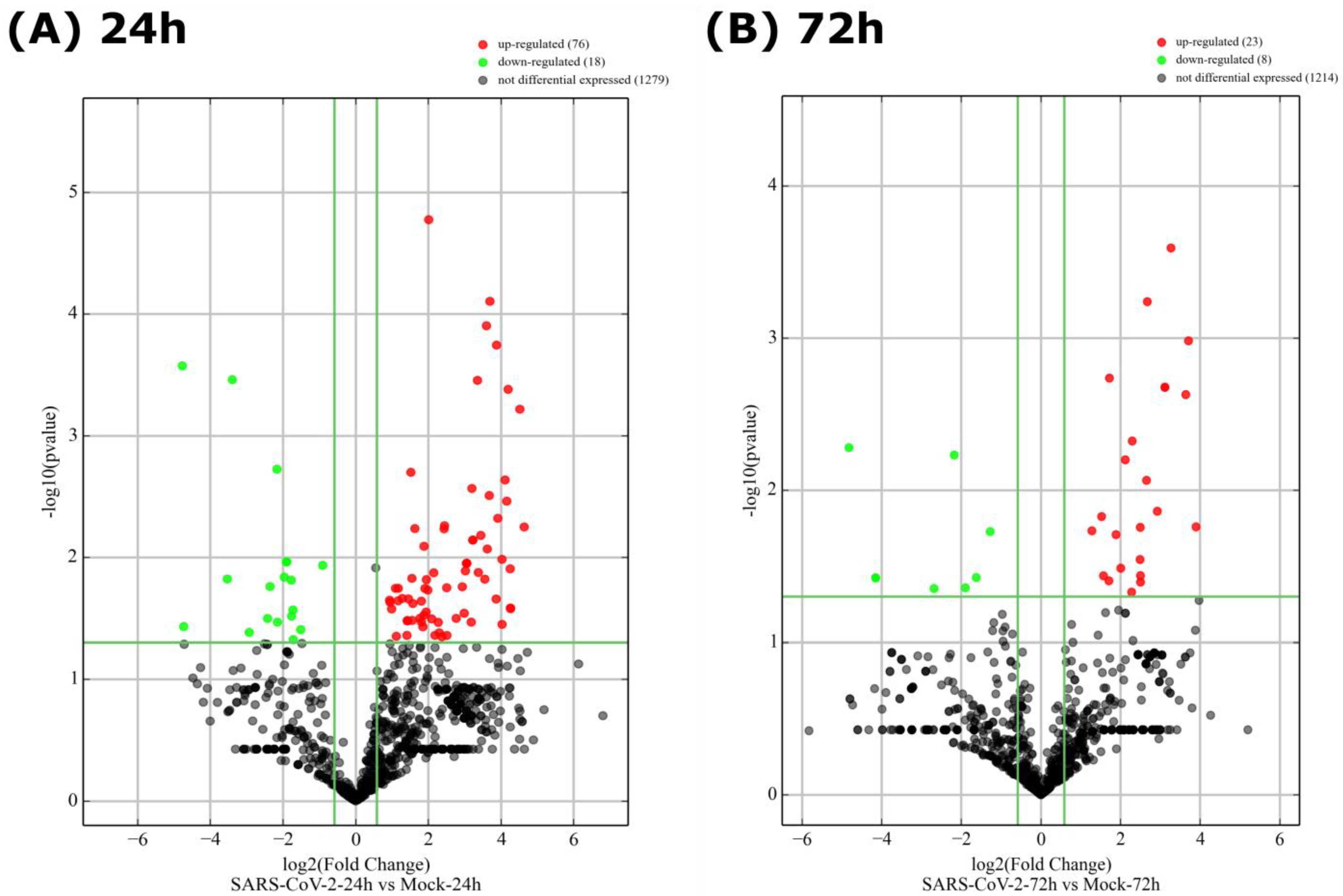
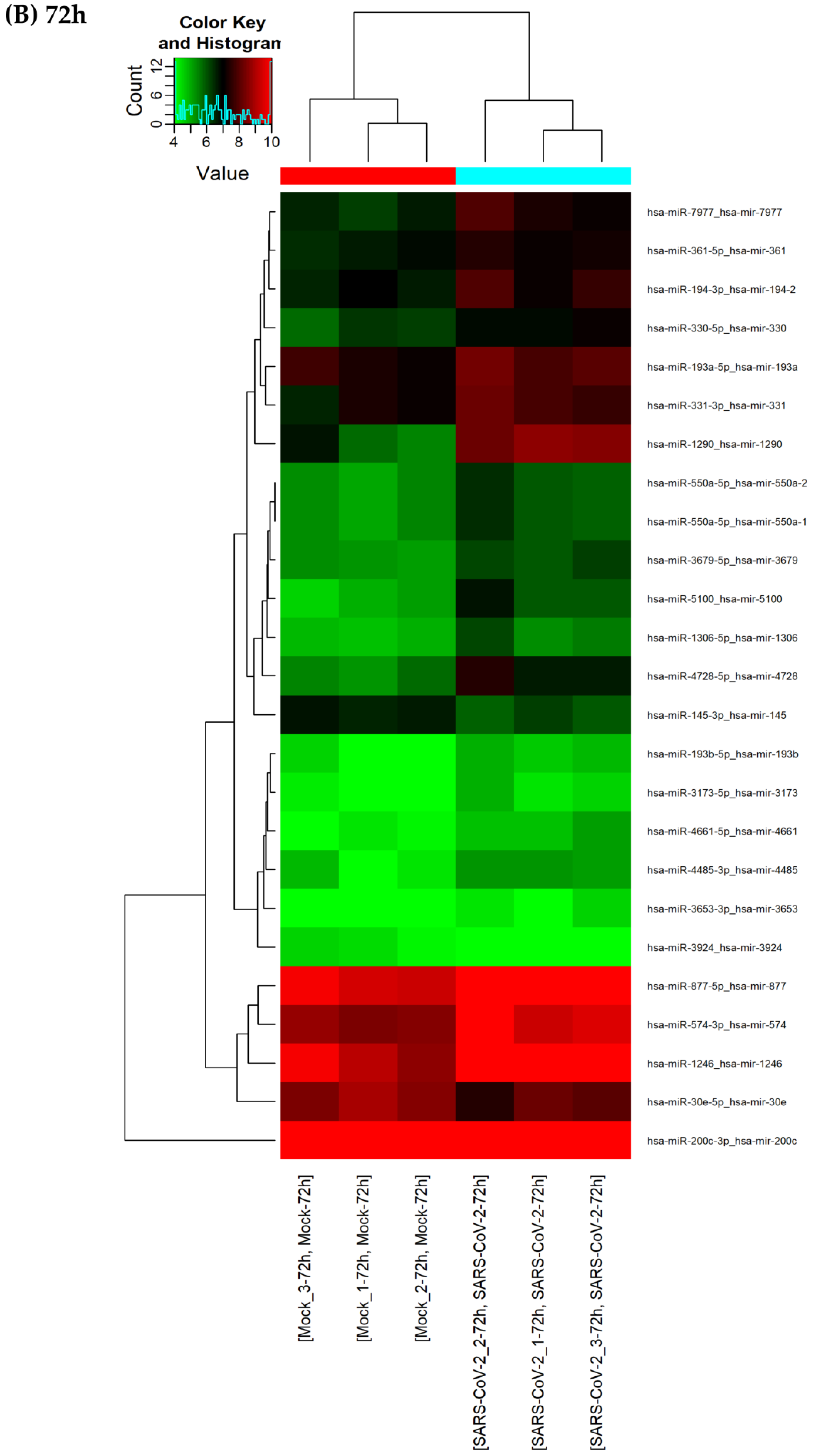
3.3. A Finite Number of miRNAs Are Differentially Expressed upon SARS-CoV-2 Infection
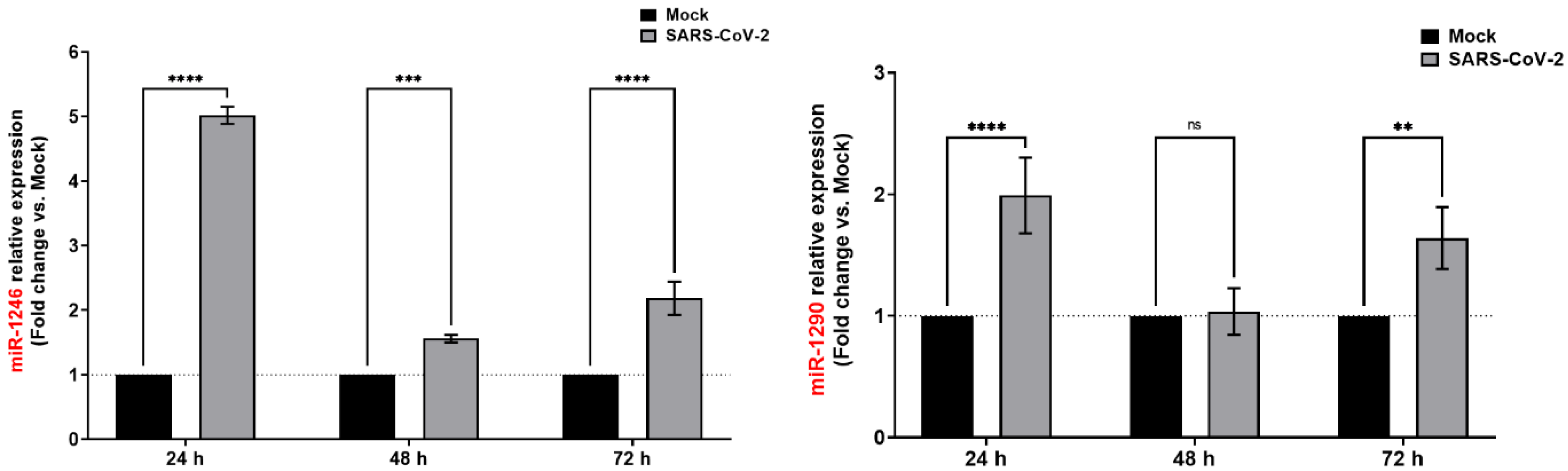
3.4. SARS-CoV-2 Infection Induces Consistent Upregulation of miR-1246 and miR-1290
3.5. miR-1246 Targets and Regulates ACE2
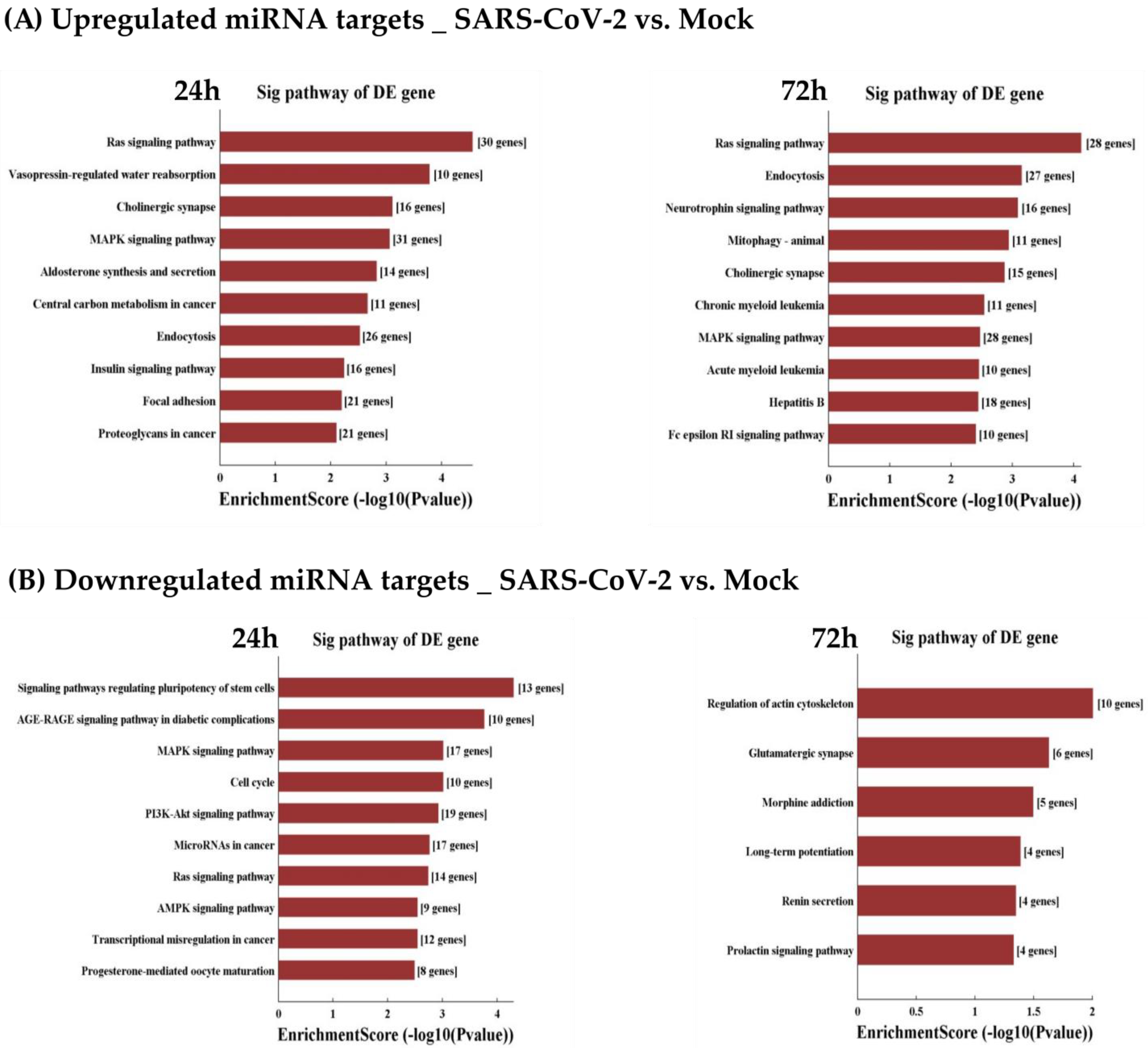
3.6. GO and KEGG Analyses of Differentially Expressed miRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- COVID-19 Map. Available online: https://coronavirus.jhu.edu/map.html (accessed on 17 May 2022).
- Mousavizadeh, L.; Ghasemi, S. Genotype and Phenotype of COVID-19: Their Roles in Pathogenesis. J. Microbiol. Immunol. Infect. 2021, 54, 159–163. [Google Scholar] [CrossRef]
- Anand, U.; Jakhmola, S.; Indari, O.; Jha, H.C.; Chen, Z.-S.; Tripathi, V.; Pérez de la Lastra, J.M. Potential Therapeutic Targets and Vaccine Development for SARS-CoV-2/COVID-19 Pandemic Management: A Review on the Recent Update. Front. Immunol. 2021, 12, 658519. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Khosroshahi, L.; Rokni, M.; Mokhtari, T.; Noorbakhsh, F. Immunology, Immunopathogenesis and Immunotherapeutics of COVID-19; an Overview. Int. Immunopharmacol. 2021, 93, 107364. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses Post-SARS: Update on Replication and Pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin Cleavage of SARS-CoV-2 Spike Promotes but Is Not Essential for Infection and Cell-Cell Fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef]
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Lee, J.Y.; Yang, J.W.; Lee, K.H.; Effenberger, M.; Szpirt, W.; Kronbichler, A.; Shin, J.I. Immunopathogenesis and Treatment of Cytokine Storm in COVID-19. Theranostics 2021, 11, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Patel, K.J.; Ranjan, K. COVID-19: Unmasking Emerging SARS-CoV-2 Variants, Vaccines and Therapeutic Strategies. Biomolecules 2021, 11, 993. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- El-Nabi, S.H.; Elhiti, M.; El-Sheekh, M. A New Approach for COVID-19 Treatment by Micro-RNA. Med. Hypotheses 2020, 143, 110203. [Google Scholar] [CrossRef]
- Zhang, S.; Amahong, K.; Sun, X.; Lian, X.; Liu, J.; Sun, H.; Lou, Y.; Zhu, F.; Qiu, Y. The MiRNA: A Small but Powerful RNA for COVID-19. Brief. Bioinform. 2021, 22, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular MiRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Marchi, R.; Sugita, B.; Centa, A.; Fonseca, A.S.; Bortoletto, S.; Fiorentin, K.; Ferreira, S.; Cavalli, L.R. The Role of MicroRNAs in Modulating SARS-CoV-2 Infection in Human Cells: A Systematic Review. Infect. Genet. Evol. 2021, 91, 104832. [Google Scholar] [CrossRef] [PubMed]
- Saulle, I.; Garziano, M.; Fenizia, C.; Cappelletti, G.; Parisi, F.; Clerici, M.; Cetin, I.; Savasi, V.; Biasin, M. MiRNA Profiling in Plasma and Placenta of SARS-CoV-2-Infected Pregnant Women. Cells 2021, 10, 1788. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The Noncoding and Coding Transcriptional Landscape of the Peripheral Immune Response in Patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J.-H. Differential MicroRNA Expression in the Peripheral Blood from Human Patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- Wang, S.; Yao, X.; Ma, S.; Ping, Y.; Fan, Y.; Sun, S.; He, Z.; Shi, Y.; Sun, L.; Xiao, S.; et al. A Single-Cell Transcriptomic Landscape of the Lungs of Patients with COVID-19. Nat. Cell Biol. 2021, 23, 1314–1328. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.T.-S.; Salmena, L. Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic Profiling of SARS-CoV-2 Infected Human Cell Lines Identifies HSP90 as Target for COVID-19 Therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef]
- Banerjee, A.; El-Sayes, N.; Budylowski, P.; Jacob, R.A.; Richard, D.; Maan, H.; Aguiar, J.A.; Demian, W.L.; Baid, K.; D’Agostino, M.R.; et al. Experimental and Natural Evidence of SARS-CoV-2-Infection-Induced Activation of Type I Interferon Responses. iScience 2021, 24, 102477. [Google Scholar] [CrossRef]
- Diallo, I.; Ho, J.; Laffont, B.; Laugier, J.; Benmoussa, A.; Lambert, M.; Husseini, Z.; Soule, G.; Kozak, R.; Kobinger, G.P.; et al. Altered MicroRNA Transcriptome in Cultured Human Liver Cells upon Infection with Ebola Virus. Int. J. Mol. Sci. 2021, 22, 3792. [Google Scholar] [CrossRef]
- Diallo, I.; Ho, J.; Lalaouna, D.; Massé, E.; Provost, P. RNA Sequencing Unveils Very Small RNAs With Potential Regulatory Functions in Bacteria. Front. Mol. Biosci. 2022, 9, 914991. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.; Guellal, S.; Ho, J.; Benmoussa, A.; Laffont, B.; Bélanger, R.; Provost, P. An Expanded Landscape of Unusually Short RNAs in 11 Samples from Six Eukaryotic Organisms. Non-Coding RNA 2022, 8, 34. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vester, B.; Wengel, J. LNA (Locked Nucleic Acid): High-Affinity Targeting of Complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef]
- Diallo, I.; Husseini, Z.; Guellal, S.; Vion, E.; Ho, J.; Kozak, R.A.; Kobinger, G.P.; Provost, P. Ebola Virus Encodes Two MicroRNAs in Huh7-Infected Cells. Int. J. Mol. Sci. 2022, 23, 5228. [Google Scholar] [CrossRef]
- Banerjee, A.; Nasir, J.A.; Budylowski, P.; Yip, L.; Aftanas, P.; Christie, N.; Ghalami, A.; Baid, K.; Raphenya, A.R.; Hirota, J.A.; et al. Isolation, Sequence, Infectivity, and Replication Kinetics of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 2054–2063. [Google Scholar] [CrossRef] [PubMed]
- McBride, R.; van Zyl, M.; Fielding, B.C. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The Cytokine Storm in COVID-19: An Overview of the Involvement of the Chemokine/Chemokine-Receptor System. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 Infection: The Role of Cytokines in COVID-19 Disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Kedzierski, L.; Linossi, E.M.; Kolesnik, T.B.; Day, E.B.; Bird, N.L.; Kile, B.T.; Belz, G.T.; Metcalf, D.; Nicola, N.A.; Kedzierska, K.; et al. Suppressor of Cytokine Signaling 4 (SOCS4) Protects against Severe Cytokine Storm and Enhances Viral Clearance during Influenza Infection. PLoS Pathog. 2014, 10, e1004134. [Google Scholar] [CrossRef]
- Kopan, R. Notch Signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a011213. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt Signal Transduction Pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Ji, W.-T.; Liu, H.J. PI3K-Akt Signaling and Viral Infection. Recent Pat. Biotechnol. 2008, 2, 218–226. [Google Scholar] [CrossRef]
- Beyerstedt, S.; Casaro, E.B.; Rangel, É.B. COVID-19: Angiotensin-Converting Enzyme 2 (ACE2) Expression and Tissue Susceptibility to SARS-CoV-2 Infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of Angiotensin-Converting Enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, D.L.; Plante, I.; Landry, P.; Barat, C.; Janelle, M.-E.; Flamand, L.; Tremblay, M.J.; Provost, P. Identification of Functional MicroRNAs Released through Asymmetrical Processing of HIV-1 TAR Element. Nucleic Acids Res. 2008, 36, 2353–2365. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, D.L.; Vigneault-Edwards, J.; Létourneau, K.; Gobeil, L.-A.; Plante, I.; Burnett, J.C.; Rossi, J.J.; Provost, P. Regulation of Host Gene Expression by HIV-1 TAR MicroRNAs. Retrovirology 2013, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, D.L.; Plante, I.; Boissonneault, V.; Ayari, C.; Provost, P. Refractoriness of Hepatitis C Virus Internal Ribosome Entry Site to Processing by Dicer in Vivo. J. Negat. Results BioMed. 2009, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Shin, O.S.; Kumar, M.; Yanagihara, R.; Song, J.-W. Hantaviruses Induce Cell Type- and Viral Species-Specific Host MicroRNA Expression Signatures. Virology 2013, 446, 217–224. [Google Scholar] [CrossRef]
- Choi, E.-J.; Kim, H.B.; Baek, Y.H.; Kim, E.-H.; Pascua, P.N.Q.; Park, S.-J.; Kwon, H.-I.; Lim, G.-J.; Kim, S.; Kim, Y.-I.; et al. Differential MicroRNA Expression Following Infection with a Mouse-Adapted, Highly Virulent Avian H5N2 Virus. BMC Microbiol. 2014, 14, 252. [Google Scholar] [CrossRef]
- Roustai Geraylow, K.; Hemmati, R.; Kadkhoda, S.; Ghafouri-Fard, S. MiRNA Expression in COVID-19. Gene Rep. 2022, 28, 101641. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- tenOever, B.R. RNA Viruses and the Host MicroRNA Machinery. Nat. Rev. Microbiol. 2013, 11, 169–180. [Google Scholar] [CrossRef]
- Backes, C.; Meese, E.; Keller, A. Specific MiRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef]
- Kucher, A.N.; Koroleva, I.A.; Zarubin, A.A.; Nazarenko, M.S. MicroRNAs as the Potential Regulators of SARS-CoV-2 Infection and Modifiers of the COVID-19 Clinical Features. Mol. Biol. 2022, 56, 29–45. [Google Scholar] [CrossRef]
- Masliah-Planchon, J.; Garinet, S.; Pasmant, E. RAS-MAPK Pathway Epigenetic Activation in Cancer: MiRNAs in Action. Oncotarget 2016, 7, 38892–38907. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS Is Regulated by the Let-7 MicroRNA Family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, J.; Gao, J.; Li, Y. MicroRNA-4728 Mediated Regulation of MAPK Oncogenic Signaling in Papillary Thyroid Carcinoma. Saudi J. Biol. Sci. 2018, 25, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Samadian, M. A Review on the Role of MiR-1246 in the Pathoetiology of Different Cancers. Front. Mol. Biosci. 2021, 8, 771835. [Google Scholar] [CrossRef]
- Lin, M.; Shi, C.; Lin, X.; Pan, J.; Shen, S.; Xu, Z.; Chen, Q. SMicroRNA-1290 Inhibits Cells Proliferation and Migration by Targeting FOXA1 in Gastric Cancer Cells. Gene 2016, 582, 137–142. [Google Scholar] [CrossRef]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 Infects Cells after Viral Entry via Clathrin-Mediated Endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Kopańska, M.; Batoryna, M.; Bartman, P.; Szczygielski, J.; Banaś-Ząbczyk, A. Disorders of the Cholinergic System in COVID-19 Era—A Review of the Latest Research. Int. J. Mol. Sci. 2022, 23, 672. [Google Scholar] [CrossRef]
- Bhutta, M.S.; Gallo, E.S.; Borenstein, R. Multifaceted Role of AMPK in Viral Infections. Cells 2021, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Khezri, M.R.; Varzandeh, R.; Ghasemnejad-Berenji, M. The Probable Role and Therapeutic Potential of the PI3K/AKT Signaling Pathway in SARS-CoV-2 Induced Coagulopathy. Cell. Mol. Biol. Lett. 2022, 27, 6. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. MiRNA Interplay: Mechanisms and Consequences in Cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef]
- Fang, Y.; Gao, F.; Hao, J.; Liu, Z. MicroRNA-1246 Mediates Lipopolysaccharide-Induced Pulmonary Endothelial Cell Apoptosis and Acute Lung Injury by Targeting Angiotensin-Converting Enzyme 2. Am. J. Transl. Res. 2017, 9, 1287–1296. [Google Scholar] [PubMed]
- Cazorla-Rivero, S.; Mura-Escorche, G.; Gonzalvo-Hernández, F.; Mayato, D.; Córdoba-Lanús, E.; Casanova, C. Circulating MiR-1246 in the Progression of Chronic Obstructive Pulmonary Disease (COPD) in Patients from the BODE Cohort. Int. J. Chronic Obstr. Pulm. Dis. 2020, 15, 2727–2737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Rostami, M.R.; Leopold, P.L.; Mezey, J.G.; O’Beirne, S.L.; Strulovici-Barel, Y.; Crystal, R.G. Expression of the SARS-CoV-2 ACE2 Receptor in the Human Airway Epithelium. Am. J. Respir. Crit. Care Med. 2020, 202, 219–229. [Google Scholar] [CrossRef]
- Wei, C.; Henderson, H.; Spradley, C.; Li, L.; Kim, I.-K.; Kumar, S.; Hong, N.; Arroliga, A.C.; Gupta, S. Circulating MiRNAs as Potential Marker for Pulmonary Hypertension. PLoS ONE 2013, 8, e64396. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.T.-A.; Khalid, Z.; Zahid, H.; Yousaf, M.A.; Shakoori, A.R. A Computational and Bioinformatic Analysis of ACE2: An Elucidation of Its Dual Role in COVID-19 Pathology and Finding Its Associated Partners as Potential Therapeutic Targets. J. Biomol. Struct. Dyn. 2022, 40, 1813–1829. [Google Scholar] [CrossRef] [PubMed]
- Branco, A.C.C.C.; Sato, M.N.; Alberca, R.W. The Possible Dual Role of the ACE2 Receptor in Asthma and Coronavirus (SARS-CoV2) Infection. Front. Cell. Infect. Microbiol. 2020, 10, 550571. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W. Mechanisms of MicroRNA-Mediated Gene Regulation in Animal Cells. Trends Genet. 2007, 23, 243–249. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Bungau, S.; Kumar, A.; Uddin, M.S.; Kumar, C.; Pal, G.; Shrivastava, K.; Zengin, G.; Arora, S. The Dual Impact of ACE2 in COVID-19 and Ironical Actions in Geriatrics and Pediatrics with Possible Therapeutic Solutions. Life Sci. 2020, 257, 118075. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, A.; Ferrara, F. Correlation between Renin-Angiotensin System and Severe Acute Respiratory Syndrome Coronavirus 2 Infection: What Do We Know? Eur. J. Pharmacol. 2020, 883, 173373. [Google Scholar] [CrossRef]
- Wiese, O.J.; Allwood, B.W.; Zemlin, A.E. COVID-19 and the Renin-Angiotensin System (RAS): A Spark That Sets the Forest Alight? Med. Hypotheses 2020, 144, 110231. [Google Scholar] [CrossRef]
- Bavishi, C.; Maddox, T.M.; Messerli, F.H. Coronavirus Disease 2019 (COVID-19) Infection and Renin Angiotensin System Blockers. JAMA Cardiol. 2020, 5, 745–747. [Google Scholar] [CrossRef]
- Pagliaro, P.; Penna, C. ACE/ACE2 Ratio: A Key Also in 2019 Coronavirus Disease (COVID-19)? Front. Med. 2020, 7, 335. [Google Scholar] [CrossRef]
- Bastolla, U.; Chambers, P.; Abia, D.; Garcia-Bermejo, M.-L.; Fresno, M. Is Covid-19 Severity Associated With ACE2 Degradation? Front. Drug Discov. 2022, 1, 789710. [Google Scholar] [CrossRef]
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Lorè, N.I.; De Lorenzo, R.; Rancoita, P.M.V.; Cugnata, F.; Agresti, A.; Benedetti, F.; Bianchi, M.E.; Bonini, C.; Capobianco, A.; Conte, C.; et al. CXCL10 Levels at Hospital Admission Predict COVID-19 Outcome: Hierarchical Assessment of 53 Putative Inflammatory Biomarkers in an Observational Study. Mol. Med. 2021, 27, 129. [Google Scholar] [CrossRef] [PubMed]
- Rubin, E.J.; Longo, D.L.; Baden, L.R. Interleukin-6 Receptor Inhibition in Covid-19—Cooling the Inflammatory Soup. N. Engl. J. Med. 2021, 384, 1564–1565. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809. [Google Scholar] [CrossRef]
- Kalil, A.C.; Mehta, A.K.; Patterson, T.F.; Erdmann, N.; Gomez, C.A.; Jain, M.K.; Wolfe, C.R.; Ruiz-Palacios, G.M.; Kline, S.; Pineda, J.R.; et al. Efficacy of Interferon Beta-1a plus Remdesivir Compared with Remdesivir Alone in Hospitalised Adults with COVID-19: A Double-Blind, Randomised, Placebo-Controlled, Phase 3 Trial. Lancet Respir. Med. 2021, 9, 1365–1376. [Google Scholar] [CrossRef]




| Mature miRNAs | Fold Change | p-Value (Unpaired) | FDR |
|---|---|---|---|
| SARS-CoV-2-24 h vs. Mock-24 h | |||
| hsa-miR-1246 | 14.550813 | 0.001936356 | 0.124553971 |
| hsa-miR-1290 | 10.151515 | 0.001616931 | 0.124553971 |
| hsa-miR-4728-5p | 2.855422 | 0.032727219 | 0.340049232 |
| hsa-miR-4745-5p | 2.552941 | 0.002060005 | 0.124553971 |
| hsa-miR-6842-3p | 2.520833 | 0.00909142 | 0.246889589 |
| hsa-miR-4286 | 2.278689 | 0.040817409 | 0.366081138 |
| hsa-miR-425-5p | 2.263918 | 0.036130815 | 0.348556098 |
| hsa-miR-29c-5p | 2.195652 | 0.047218269 | 0.408413103 |
| hsa-miR-3187-3p | 2.190476 | 0.018900103 | 0.293985775 |
| hsa-miR-486-5p | 2.147124 | 0.031260241 | 0.340049232 |
| SARS-CoV-2-72 h vs. Mock-72 h | |||
| hsa-miR-1246 | 6.234131 | 0.000859555 | 0.465454769 |
| hsa-miR-1290 | 5.393035 | 0.000599562 | 0.465454769 |
| hsa-miR-4728-5p | 2.768657 | 0.032469057 | 0.605339181 |
| hsa-miR-5100 | 2.522222 | 0.038432487 | 0.605339181 |
| hsa-miR-7977 | 2.098113 | 0.043036275 | 0.605339181 |
| hsa-miR-574-3p | 2.036113 | 0.015325686 | 0.605339181 |
| hsa-miR-194-3p | 1.897764 | 0.048234042 | 0.605339181 |
| hsa-miR-877-5p | 1.861196 | 0.007521499 | 0.605339181 |
| hsa-miR-1306-5p | 1.860465 | 0.046973589 | 0.605339181 |
| Mature miRNA | Fold Change | p-Value (Unpaired) | FDR |
|---|---|---|---|
| SARS-CoV-2-24 h vs. Mock-24 h | |||
| hsa-miR-196b-5p | 0.374473 | 0.036770814 | 0.351774119 |
| hsa-miR-196a-5p | 0.521818 | 0.01962821 | 0.293985775 |
| hsa-miR-196a-5p | 0.540311 | 0.023390933 | 0.308652771 |
| hsa-let-7g-5p | 0.633044 | 0.008259531 | 0.246889589 |
| hsa-let-7a-3p | 0.643836 | 0.000312666 | 0.05127724 |
| SARS-CoV-2-72 h vs. Mock-72 h | |||
| hsa-miR-3924 | 0.587302 | 0.011169221 | 0.605339181 |
| hsa-miR-30e-5p | 0.609796 | 0.040388915 | 0.605339181 |
| hsa-miR-145-3p | 0.642384 | 0.003165844 | 0.605339181 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diallo, I.; Jacob, R.A.; Vion, E.; Kozak, R.A.; Mossman, K.; Provost, P. Altered microRNA Transcriptome in Cultured Human Airway Cells upon Infection with SARS-CoV-2. Viruses 2023, 15, 496. https://doi.org/10.3390/v15020496
Diallo I, Jacob RA, Vion E, Kozak RA, Mossman K, Provost P. Altered microRNA Transcriptome in Cultured Human Airway Cells upon Infection with SARS-CoV-2. Viruses. 2023; 15(2):496. https://doi.org/10.3390/v15020496
Chicago/Turabian StyleDiallo, Idrissa, Rajesh Abraham Jacob, Elodie Vion, Robert A. Kozak, Karen Mossman, and Patrick Provost. 2023. "Altered microRNA Transcriptome in Cultured Human Airway Cells upon Infection with SARS-CoV-2" Viruses 15, no. 2: 496. https://doi.org/10.3390/v15020496







