Plant Virus-Derived Vectors for Plant Genome Engineering
Abstract
:1. Introduction
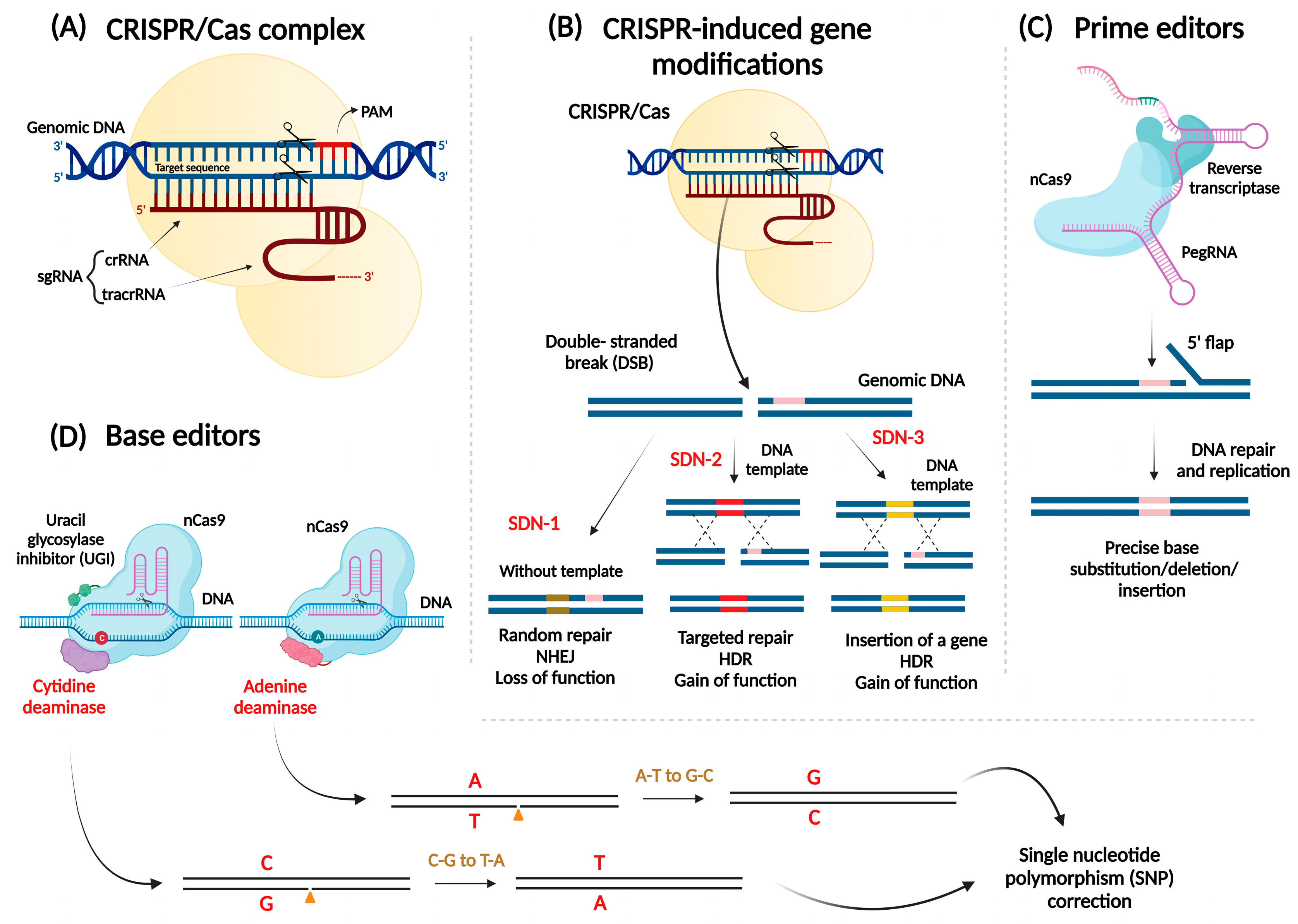
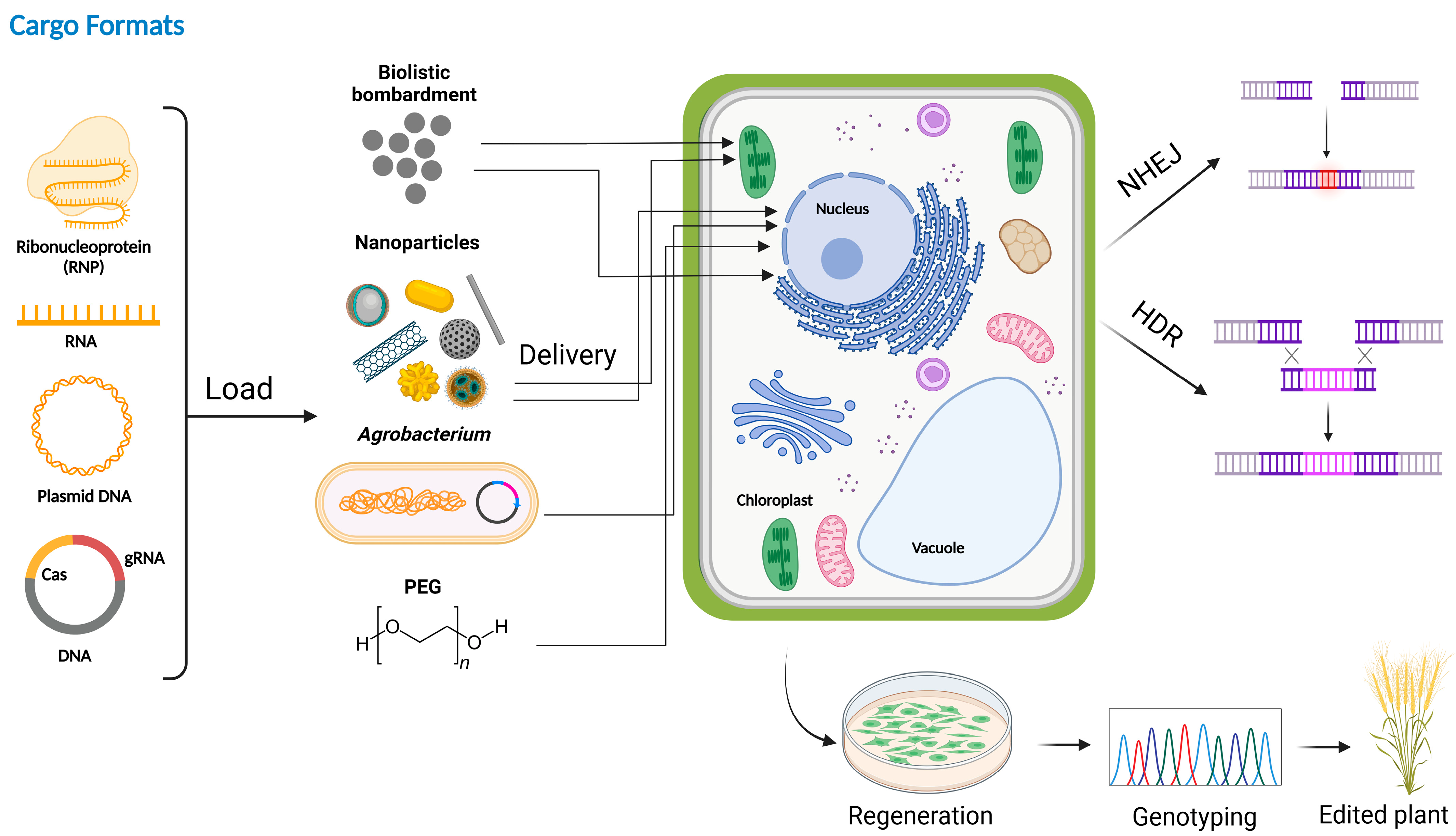
2. Ways to Deliver CRISPR/Cas Reagents to Plants
2.1. Agrobacterium-Mediated Transformation for Genome Engineering
2.2. Biolistic Transformation for Genome Engineering
2.3. Nano-Particle Based Delivery Systems for Genome Engineering
2.4. PEG-Mediated Reagent Delivery for Plant Genome Engineering
| Transformation Method | Species | Target Genes | CRISPR System | Plant Material | Editing Efficiency | References |
|---|---|---|---|---|---|---|
| Protoplast transformation | ||||||
| PEG | Arabidopsis thaliana | Allene oxide cyclase (AOC) | Cas9 | protoplast | 16% | [70] |
| BRASSINOSTEROID INSENSITIVE 1 (BRI1) | Cas9 with two gRNAs simultaneously | protoplast | 54–71% | |||
| PEG | Brassica napus cv. Topaz | Phytoene desaturase (PDS) | Cas9 | Protoplast | 0 | [74] |
| FRIGIDA (FRI) | 0 | |||||
| PEG | Brassica oletacea var. capitata f. alba) | PDS | Cas9 | Protoplast | 0.14–1.33% | [74] |
| FRI | 0.09–2.25% | |||||
| PEG | Brassica oletacea var. capitata f. alba) | GIGANTEA (GI) | Cas9 | Protoplast | 2% | [75] |
| PEG | Hot pepper (Capsicum annuum cv. CM334) | Mildew locus O 2 (MLO2) | Cas9 | Callus and protoplast | 0.2% and 17.6% | [76] |
| Sweet pepper (C. annuum cv. Dempsey) | Leaf protoplast | 0.5–11.3% | ||||
| PEG | Rice (Oryza sativa) | P450 | Cas9 | Protoplast | 19% | [70] |
| DWDI | 8.4% | |||||
| PEG | Wild tobacco (N. attenuata) | Phytochrome B, PHYB | Cas9 | Protoplast | 44% | [70] |
| PEG | Garden petunia (Petunia × hybrida) | Nitrate reductase (NR) | Cas9 Four gRNAs | Protoplast | 5.30–17.83% | [77] |
| PEG | Rice (O. sativa cv. Nipponbare) | DsRed2 | Cas9 | Zygotes produced by gamete fusion | 25% | [71] |
| DROOPING LEAF (DL) | 13.6–14.3% | |||||
| GRAIN WIDTH 7 (GW7) | 21.4% | |||||
| GENERATIVE CELL SPECIFIC-1 (GCS1) | 64.3% | |||||
| Agrobacterium-mediated and Particle bombardment-mediated delivery | ||||||
| Particle bombardment | Rice (O. sativa cv. Nipponbare) | PDS | Cas9 with the plasmid encoding hygromycin phosphotransferase (hpt) | Scutellum derived embryos | 3.6% | [78] |
| HiFi Cas9 with the plasmid encoding hpt | 8.8% | |||||
| Cas9 D10A with two gRNAs and the plasmid encoding hpt | 0 | |||||
| Cas9 with two gRNAs and the plasmid encoding hygromycin phosphotransferase (hpt) | 62.9% | [79] | ||||
| Particle bombardment | Bread wheat (Triticum aestivum cv. Kenong 199) | Grain width and weight 2 (GW2) | Cas9 | Immature embryo | 2.2% (TaGW2-B1) 4.4% (TaGW2-D1) | [80] |
| Particle bombardment | Wild tobacco (N. tabacum cv. Bright Yellow 2) | ppor-RFP | Cas9 | BY2 cells | 3% | [81] |
| Particle bombardment | Bread wheat (T. aestivum cv. YZ814) | GW2 | Cas9 | Immature embryo | 1.3% | [80] |
| GASR7 | 1.8% | |||||
| Particle bombardment | Maize (Zea mays) | Male fertility gene (MS45) | Cas9 RNP with DNA vectors encoding “helper genes” cell division-promoting transcription factors (maize ovule developmental protein 2 [ODP2] and maize Wuschel [WUS]) and selectable and visible marker genes (MOPAT-DSRED fusion) | Immature embryo | 47% (28% monoallelic mutations; 19% biallelic mutations) | [58] |
| Acetolactate synthase (ALS2) | Cas9 RNP with DNA vectors encoding helper genes; 127 nt single-stranded DNA donor | ~2–2.5% (all monoallelic mutations) | ||||
| MS45 | Cas9 RNP only | 4.0% (3.1% biallelic mutations) | ||||
| Male fertility gene (MS26) | Cas9 RNP only | 2.4% (0.3% biallelic mutations) | ||||
| Liguleless 1 (LIG) | Cas9 RNP only | 9.7% (0.9% biallelic mutations) | ||||
| Agrobacterium-mediated | A. thaliana | AtPDS3 | CRISPR components | Leaf | 37.7–38.5% | [82] |
| Tobacco (N. benthamiana) | NbPDS | 1.8–2.4% | ||||
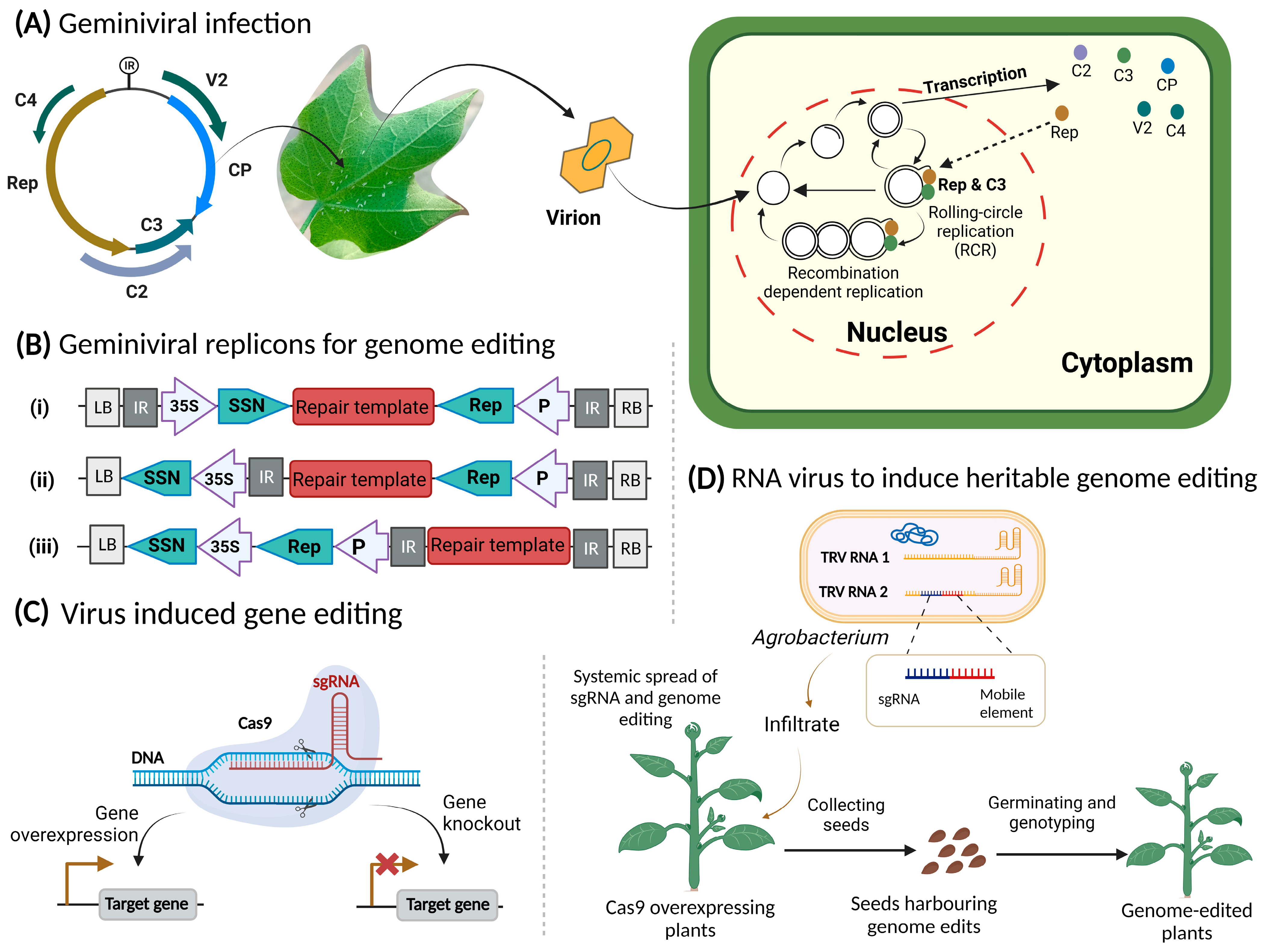
3. Plant Viruses and Their Role in CRISPR Reagents Delivery
3.1. Genome Editing by Geminiviruses: How They Can Help?
3.2. RNA Viruses a Potential Delivery System In-Planta
| Virus Type | gRNA | Nucleases Type | Plant Species | Mutation Heritability | Reference |
|---|---|---|---|---|---|
| DNA viruses | |||||
| CaLCuV | AtU6-gRNA | _ | SpCas9-overproducing tobacco (N. benthamiana) | No | [111] |
| WDV | TaU6-gRNA | SpCas9 | Wheat (T. aestivum) | No | [93] |
| OsU6-gRNA | SpCas9 | SpCas9-overproducing rice (O. sativa) | No | [114] | |
| BeYDV | AtU6-gRNA | ZFN, TALEN, SpCas9 | Tobacco (N. tabacum) | No | [91] |
| AtU6-gRNA | TALEN, SpCas9 | Tomato cv. MicroTom | No | [94] | |
| AtU6-gRNA | TALEN, SpCas9 | Potato | No | [96] | |
| AtU6-gRNA | SpCas9 | Potato | No | [95] | |
| AtU6-gRNA | SpCas9 | Tomato cv. MicroTom | No | [115] | |
| Positive (+) strand RNA virus | |||||
| TRV | _ | ZFN | Petunia (Petunia hybrida), Tobacco (N. Tabacum) | No | [106] |
| _ | Meganucleases | Tobacco (N. alata) | Yes, Low frequency | [107] | |
| PEBV-gRNA | _ | SpCas9-overproducing tobacco (N. benthamiana) | Yes, Low frequency | [101] | |
| PEBV-gRNA | _ | SpCas9-overproducing Arabidopsis thaliana, Tobacco (N. benthamiana) | No | [108] | |
| PEBV-gRNA-FT | _ | SpCas9-overproducing tobacco (N. benthamiana) | Yes, High frequency | [112] | |
| PVX | BMV-gRNA-tRNA | _ | SpCas9-overproducing tobacco (N. benthamiana) | No | [116] |
| PVX-gRNA | SpCas9/AID | N. benthamiana | No | [117] | |
| PEBV | PEBV-gRNA | _ | SpCas9-overproducing A. thaliana, Tobacco (N. benthamiana) | No | [108] |
| FoMV | FoMV-gRNA | _ | SpCas9-overproducing A. thaliana, Maize (Zea mays), Setaria viridis | No | [103] |
| FoMV | AtU6-gRNA | SpCas9 | N. benthamiana | No | [118] |
| TMV | TMV-gRNA-ribozyme | _ | N. benthamiana | No | [102] |
| BSMV | BSMV-gRNA | - | SpCas9-overproducing tobacco (N. benthamiana), Maize (Zea mays), wheat (T. aestivum) | No | [109] |
| BNYVV | p31-gRNA | _ | SpCas9 overproducing tobacco (N. benthamiana) | No | [104] |
| ToMV | AtU6-gRNA | Split-SaCas9 | N. benthamiana | No | [119] |
| Negative (−) strand RNA virus | |||||
| BYSMV | BYSMV-gRNA | SpCas9 | Tobacco N. benthamiana | No | [120] |
| SYNV | SYNV-gRNA-tRNA | SpCas9 | Tobacco N. benthamiana | No | [111] |
4. Pros and Cons of Viral Delivery Systems
5. Future Outlook
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Voytas, D.F.; Gao, C. Precision genome engineering and agriculture: Opportunities and regulatory challenges. PLoS Biol. 2014, 12, e1001877. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Gao, C.; Qiu, J.-L. Progress and prospects in plant genome editing. Nat. Plants. 2017, 3, 17107. [Google Scholar] [CrossRef] [PubMed]
- Gionfriddo, M.; De Gara, L.; Loreto, F. Directed evolution of plant processes: Towards a green (r) evolution? Trends Plant Sci. 2019, 24, 999–1007. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end joining pathway. Annu. Rev. Biochem. 2010, 79, 181. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, C.; Yang, Y.; Zhao, S.; Kang, G.; He, X.; Song, J.; Yang, J. Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant 2020, 13, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Daròs, J.A. Tools and targets: The dual role of plant viruses in CRISPR–Cas genome editing. Plant Genome. 2009; early view. [Google Scholar]
- Rybicki, E.P. Plant-produced vaccines: Promise and reality. Drug Discov. Today 2009, 14, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.-S.; Hammond-Kosack, K.E.; Kanyuka, K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: Virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 2012, 160, 582–590. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-Faraj, A.; Piatek, M.; Mahfouz, M.M. Activity and specificity of TRV-mediated gene editing in plants. Plant Signal. Behav. 2015, 10, e1044191. [Google Scholar] [CrossRef]
- Ascencio-Ibánez, J.T.; Sozzani, R.; Lee, T.-J.; Chu, T.-M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef]
- Hooykaas, P.J.; Klapwijk, P.; Nuti, M.; Schilperoort, R.; Rörsch, A. Transfer of the Agrobacterium tumefaciens Ti plasmid to avirulent agrobacteria and to Rhizobium ex planta. Microbiology 1977, 98, 477–484. [Google Scholar] [CrossRef]
- Klein, T.M.; Harper, E.C.; Svab, Z.; Sanford, J.C.; Fromm, M.E.; Maliga, P. Stable genetic transformation of intact Nicotiana cells by the particle bombardment process. Proc. Natl. Acad. Sci. USA 1988, 85, 8502–8505. [Google Scholar] [CrossRef]
- Molina-Risco, M.; Ibarra, O.; Faion-Molina, M.; Kim, B.; Septiningsih, E.M.; Thomson, M.J. Optimizing Agrobacterium-mediated transformation and CRISPR-Cas9 gene editing in the tropical japonica rice variety presidio. Int. J. Mol. Sci. 2021, 22, 10909. [Google Scholar] [CrossRef]
- Ghogare, R.; Ludwig, Y.; Bueno, G.M.; Slamet-Loedin, I.H.; Dhingra, A. Genome editing reagent delivery in plants. Transgenic Res. 2021, 30, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Laforest, L.C.; Nadakuduti, S.S. Advances in delivery mechanisms of CRISPR gene-editing reagents in plants. Front Genome Ed. 2022, 4, 830178. [Google Scholar] [CrossRef]
- Kluepfel, D.; McClean, A.; Aradhya, M.; Moersfelder, J. Identification of Juglans wild relatives resistant to crown gall caused by Agrobacterium tumefaciens. In Proceedings of the II International Symposium on Wild Relatives of Subtropical and Temperate Fruit and Nut Crops 1074, Baku, Azerbaijan, 7–12 April 2014; pp. 87–94. [Google Scholar]
- Gohlke, J.; Deeken, R. Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 2014, 5, 155. [Google Scholar] [CrossRef]
- Hoekema, A.; Hirsch, P.R.; Hooykaas, P.J.; Schilperoort, R.A. A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 1983, 303, 179–180. [Google Scholar] [CrossRef]
- Nabi, N.; Hafsa, A.B.; Gaillard, V.; Nesme, X.; Chaouachi, M.; Vial, L. Evolutionary classification of tumor-and root-inducing plasmids based on T-DNAs and virulence regions. Mol. Phylogenet. Evol. 2022, 169, 107388. [Google Scholar] [CrossRef]
- Tzfira, T.; Citovsky, V. From host recognition to T-DNA integration: The function of bacterial and plant genes in the Agrobacterium–plant cell interaction. Mol. Plant Pathol. 2000, 1, 201–212. [Google Scholar] [CrossRef]
- Nester, E.W. Agrobacterium: Nature’s genetic engineer. Front Plant Sci. 2015, 5, 730. [Google Scholar] [CrossRef]
- Shreni Agrawal, E.R. A Review: Agrobacterium-mediated gene transformation to increase plant productivity. J. Phytopharm. 2022, 11, 111–117. [Google Scholar] [CrossRef]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P. Advancing crop transformation in the era of genome editing. Plant Cell. 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed]
- Danilo, B.; Perrot, L.; Mara, K.; Botton, E.; Nogué, F.; Mazier, M. Efficient and transgene-free gene targeting using Agrobacterium-mediated delivery of the CRISPR/Cas9 system in tomato. Plant Cell Rep. 2019, 38, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Ozyigit, I.I.; Dogan, I.; Kaya, Y.; Bajrovic, K.; Gozukirmizi, N. Cotton biotechnology: An efficient gene transfer protocol via agrobacterium tumefaciens for a greater transgenic recovery. J. Nat. Fibers 2022, 19, 1–15. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Sandhya, D.; Jogam, P.; Venkatapuram, A.K.; Savitikadi, P.; Peddaboina, V.; Allini, V.R.; Abbagani, S. Highly efficient Agrobacterium-mediated transformation and plant regeneration system for genome engineering in tomato. Saudi J. Biol. Sci. 2022, 29, 103292. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, K.; Finley, T.; Chappell, H.; Veena, V.; Wang, K. An improved Agrobacterium-mediated transformation and genome-editing method for maize inbred B104 using a ternary vector system and immature embryos. Front Plant Sci. 2022, 13, 860971. [Google Scholar] [CrossRef]
- Xu, H.; Guo, Y.; Qiu, L.; Ran, Y. Progress in Soybean Genetic Transformation Over the Last Decade. Front Plant Sci. 2022, 13, 900318. [Google Scholar] [CrossRef]
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 2017, 15, 257–268. [Google Scholar] [CrossRef]
- Toinga-Villafuerte, S.; Janga, M.R.; Isabel Vales, M.; Rathore, K.S. Green fluorescent protein gene as a tool to examine the efficacy of Agrobacterium-delivered CRISPR/Cas9 reagents to generate targeted mutations in the potato genome. Plant Cell 2022, 150, 587–598. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, G.; Cheng, C.; Lei, L.; Sun, J.; Xu, Y.; Deng, C.; Dai, Z.; Yang, Z.; Chen, X. Establishment of an Agrobacterium-mediated genetic transformation and CRISPR/Cas9-mediated targeted mutagenesis in Hemp (Cannabis sativa L.). Plant Biotechnol. J. 2021, 19, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Li, Q.; Peng, W.; Zhang, Z.; Chu, P.; Guo, S.; Fan, Y.; Lyu, S. AtGCS promoter-driven clustered regularly interspaced short palindromic repeats/Cas9 highly efficiently generates homozygous/biallelic mutations in the transformed roots by Agrobacterium rhizogenes–mediated transformation. Front Plant Sci. 2022, 13, 952428. [Google Scholar] [CrossRef]
- Kharb, P.; Chaudhary, R.; Tuteja, N.; Kaushik, P. A Genotype-Independent, Simple, Effective and Efficient in Planta Agrobacterium-Mediated Genetic Transformation Protocol. Methods Protoc. 2022, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Aliu, E.; Lee, K.; Wang, K. CRISPR RNA-guided integrase enables high-efficiency targeted genome engineering in Agrobacterium tumefaciens. Plant Biotechnol. J. 2022, 20, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, S.D.; Karimi, M.; Impens, L.; Van Lerberge, E.; Coussens, G.; Aesaert, S.; Rombaut, D.; Holtappels, D.; Ibrahim, H.M.; Van Montagu, M. Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc. Natl. Acad. Sci. USA 2021, 118, e2013338118. [Google Scholar] [CrossRef]
- Aregawi, K.; Shen, J.; Pierroz, G.; Sharma, M.K.; Dahlberg, J.; Owiti, J.; Lemaux, P.G. Morphogene-assisted transformation of Sorghum bicolor allows more efficient genome editing. Plant Biotechnol. J. 2022, 20, 748–760. [Google Scholar] [CrossRef]
- Peterson, D.; Barone, P.; Lenderts, B.; Schwartz, C.; Feigenbutz, L.; St. Clair, G.; Jones, S.; Svitashev, S. Advances in Agrobacterium transformation and vector design result in high-frequency targeted gene insertion in maize. Plant Biotechnol. J. 2021, 19, 2000–2010. [Google Scholar] [CrossRef] [PubMed]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020, 10, 20155. [Google Scholar] [CrossRef] [PubMed]
- Kohli, A.; Twyman, R.M.; Abranches, R.; Wegel, E.; Stoger, E.; Christou, P. Transgene integration, organization and interaction in plants. Plant Mol. Biol. 2003, 52, 247–258. [Google Scholar] [CrossRef]
- Oltmanns, H.; Frame, B.; Lee, L.-Y.; Johnson, S.; Li, B.; Wang, K.; Gelvin, S.B. Generation of backbone-free, low transgene copy plants by launching T-DNA from the Agrobacterium chromosome. Plant Physiol. 2010, 152, 1158–1166. [Google Scholar] [CrossRef]
- Tsatsakis, A.M.; Nawaz, M.A.; Kouretas, D.; Balias, G.; Savolainen, K.; Tutelyan, V.A.; Golokhvast, K.S.; Lee, J.D.; Yang, S.H.; Chung, G. Environmental impacts of genetically modified plants: A review. Environ. Res. 2017, 156, 818–833. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Verma, S.; Sahoo, R.K.; Raveendar, S.; Reddy, I. Recent advances in development of marker-free transgenic plants: Regulation and biosafety concern. J. Biosci. 2012, 37, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Barton, I.S.; Fuqua, C.; Platt, T.G. Ecological and evolutionary dynamics of a model facultative pathogen: Agrobacterium and crown gall disease of plants. Environ. Microbiol. 2018, 20, 16–29. [Google Scholar] [CrossRef]
- Kuraya, Y.; Ohta, S.; Fukuda, M.; Hiei, Y.; Murai, N.; Hamada, K.; Ueki, J.; Imaseki, H.; Komari, T. Suppression of transfer of non-T-DNA ‘vector backbone’sequences by multiple left border repeats in vectors for transformation of higher plants mediated by Agrobacterium tumefaciens. Mol. Breed. 2004, 14, 309–320. [Google Scholar] [CrossRef]
- Sharma, K.K.; Anjaiah, V. An efficient method for the production of transgenic plants of peanut (Arachis hypogaea L.) through Agrobacterium tumefaciens-mediated genetic transformation. Plant Sci. 2000, 159, 7–19. [Google Scholar] [CrossRef]
- Bhatnagar, M.; Prasad, K.; Bhatnagar-Mathur, P.; Lakshmi Narasu, M.; Waliyar, F.; Sharma, K.K. An efficient method for the production of marker-free transgenic plants of peanut (Arachis hypogaea L.). Plant Cell Rep. 2010, 29, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Zhai, C.; Li, H.; Li, J.; Mei, W.; Gui, H.; Ni, D.; Song, F.; Li, L.; Zhang, W. An efficient and high-throughput protocol for Agrobacterium-mediated transformation based on phosphomannose isomerase positive selection in Japonica rice (Oryza sativa L.). Plant Cell Rep. 2012, 31, 1611–1624. [Google Scholar] [CrossRef]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Fact. 2017, 16, 168. [Google Scholar] [CrossRef] [PubMed]
- Ueki, S.; Lacroix, B.; Krichevsky, A.; Lazarowitz, S.G.; Citovsky, V. Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat. Protoc. 2009, 4, 71–77. [Google Scholar] [CrossRef]
- Klein, T.M.; Wolf, E.D.; Wu, R.; Sanford, J.C. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 1987, 327, 70–73. [Google Scholar] [CrossRef]
- Hamada, H.; Liu, Y.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 2018, 8, 14422. [Google Scholar] [CrossRef]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Mark Cigan, A. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [CrossRef]
- Nagahara, S.; Higashiyama, T.; Mizuta, Y. Detection of a biolistic delivery of fluorescent markers and CRISPR/Cas9 to the pollen tube. Plant Reprod. 2021, 34, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Eggenberger, A.L.; Lee, K.; Liu, F.; Kang, M.; Drent, M.; Ruba, A.; Kirscht, T.; Wang, K.; Jiang, S. An improved biolistic delivery and analysis method for evaluation of DNA and CRISPR-Cas delivery efficacy in plant tissue. Sci. Rep. 2021, 11, 7695. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N. DNA-free genome editing methods for targeted crop improvement. Plant Cell Rep. 2016, 35, 1469–1474. [Google Scholar] [CrossRef]
- Chilcoat, D.; Liu, Z.-B.; Sander, J. Use of CRISPR/Cas9 for crop improvement in maize and soybean. Prog. Mol. Biol. Transl. Sci. 2017, 149, 27–46. [Google Scholar]
- Sanford, J.C. Biolistic plant transformation. Physiol. Plant. 1990, 79, 206–209. [Google Scholar] [CrossRef]
- Demirer, G.S.; Silva, T.N.; Jackson, C.T.; Thomas, J.B.; Ehrhardt, D.W.; Rhee, S.Y.; Mortimer, J.C.; Landry, M.P. Nanotechnology to advance CRISPR–Cas genetic engineering of plants. Nat. Nanotechnol. 2021, 16, 243–250. [Google Scholar] [CrossRef]
- Yan, Y.; Zhu, X.; Yu, Y.; Li, C.; Zhang, Z.; Wang, F. Nanotechnology strategies for plant genetic engineering. Adv. Mater. 2022, 34, 2106945. [Google Scholar] [CrossRef] [PubMed]
- Alghuthaymi, M.A.; Ahmad, A.; Khan, Z.; Khan, S.H.; Ahmed, F.K.; Faiz, S.; Nepovimova, E.; Kuča, K.; Abd-Elsalam, K.A. Exosome/liposome-like nanoparticles: New carriers for crispr genome editing in plants. Int. J. Mol. Sci. 2021, 22, 7456. [Google Scholar] [CrossRef] [PubMed]
- Zhi, H.; Zhou, S.; Pan, W.; Shang, Y.; Zeng, Z.; Zhang, H. The Promising Nanovectors for Gene Delivery in Plant Genome Engineering. Int. J. Mol. Sci. 2022, 23, 8501. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, L.M.; Kaur, P.; Stanton, D.; Grosser, J.W.; Dutt, M. A cationic lipid mediated CRISPR/Cas9 technique for the production of stable genome edited citrus plants. Plant Methods 2022, 18, 33. [Google Scholar] [CrossRef]
- Sandhya, D.; Jogam, P.; Allini, V.R.; Abbagani, S.; Alok, A. The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Eng. Biotechnol. 2020, 18, 25. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Toda, E.; Koiso, N.; Takebayashi, A.; Ichikawa, M.; Kiba, T.; Osakabe, K.; Osakabe, Y.; Sakakibara, H.; Kato, N.; Okamoto, T. An efficient DNA-and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 2019, 5, 363–368. [Google Scholar] [CrossRef]
- González, M.N.; Massa, G.A.; Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Storani, L.; Décima Oneto, C.A.; Hofvander, P.; Feingold, S.E. Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front Plant Sci. 2020, 10, 1649. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17018. [Google Scholar] [CrossRef]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front Plant Sci. 2018, 9, 1594. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, S.; Jeong, Y.J.; Lee, S.B.; Pyun, J.W.; Kim, S.; Kim, T.H.; Kim, S.W.; Jeong, J.C.; Kim, C.Y. DNA-free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol. Rep. 2019, 13, 483–489. [Google Scholar] [CrossRef]
- Kim, H.; Choi, J. A robust and practical CRISPR/crRNA screening system for soybean cultivar editing using LbCpf1 ribonucleoproteins. Plant Cell Rep. 2021, 40, 1059–1070. [Google Scholar] [CrossRef]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.-M.; Kim, D.H.; Kim, J.-S.; Bae, S.; Lee, G.-J. Site-directed mutagenesis in Petunia× hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Banakar, R.; Schubert, M.; Collingwood, M.; Vakulskas, C.; Eggenberger, A.L.; Wang, K. Comparison of CRISPR-Cas9/Cas12a ribonucleoprotein complexes for genome editing efficiency in the rice phytoene desaturase (OsPDS) gene. Rice 2020, 13, 4. [Google Scholar] [CrossRef]
- Banakar, R.; Eggenberger, A.L.; Lee, K.; Wright, D.A.; Murugan, K.; Zarecor, S.; Lawrence-Dill, C.J.; Sashital, D.G.; Wang, K. High-frequency random DNA insertions upon co-delivery of CRISPR-Cas9 ribonucleoprotein and selectable marker plasmid in rice. Sci. Rep. 2019, 9, 19902. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Liu, W.; Rudis, M.R.; Cheplick, M.H.; Millwood, R.J.; Yang, J.-P.; Ondzighi-Assoume, C.A.; Montgomery, G.A.; Burris, K.P.; Mazarei, M.; Chesnut, J.D. Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 2020, 39, 245–257. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE 2015, 10, e0136064. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.-B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef]
- Xu, C.L.; Ruan, M.Z.; Mahajan, V.B.; Tsang, S.H. Viral delivery systems for CRISPR. Viruses 2019, 11, 28. [Google Scholar] [CrossRef]
- Mahmood, M.; Naqvi, R.; Irfan, N.; Amin, I.; Mansoor, S. First report of Cotton leaf curl Multan virus infecting Millettia pinnata in Pakistan. New Dis. Rep. 2022, 46, e12116. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Ahmed, N.; Hussain, S.; Muntaha, S.T.; Amin, I.; Mansoor, S. Dominance of Asia II 1 species of Bemisia tabaci in Pakistan and beyond. Sci. Rep. 2022, 12, 1528. [Google Scholar] [CrossRef]
- Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Evolution of geminiviruses and their satellites. FEBS Lett. 2009, 583, 1825–1832. [Google Scholar] [CrossRef]
- Ahmed, N.; Mahmood, M.A.; Amin, I.; Mansoor, S. Geminiviruses also encode small proteins with specific functions. Trends Microbiol. 2021, 29, 1052–1054. [Google Scholar] [CrossRef]
- Lozano-Durán, R. Geminiviruses for biotechnology: The art of parasite taming. N. Phytol. 2016, 210, 58–64. [Google Scholar] [CrossRef]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef]
- Richter, K.S.; Serra, H.; White, C.I.; Jeske, H. The recombination mediator RAD51D promotes geminiviral infection. Virology 2016, 493, 113–127. [Google Scholar] [CrossRef]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE 2015, 10, e0144591. [Google Scholar] [CrossRef]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Cody, W.B.; Scholthof, H.B. Plant virus vectors 3.0: Transitioning into synthetic genomics. Annu. Rev. Phytopathol. 2019, 57, 211–230. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Li, X.; Zhang, R.; Li, J. Virus-Induced Gene Editing and Its Applications in Plants. Int. J. Mol. Sci. 2022, 23, 10202. [Google Scholar] [CrossRef]
- Oh, Y.; Kim, H.; Kim, S.-G. Virus-induced plant genome editing. Curr. Opin. Plant Biol. 2021, 60, 101992. [Google Scholar] [CrossRef]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.L.; Liu, Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-Faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef]
- Cody, W.B.; Scholthof, H.B.; Mirkov, T.E. Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant Physiol. 2017, 175, 23–35. [Google Scholar] [CrossRef]
- Mei, Y.; Beernink, B.M.; Ellison, E.E.; Konečná, E.; Neelakandan, A.K.; Voytas, D.F.; Whitham, S.A. Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct 2019, 3, e00181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Zhang, C.; Liu, J.Y.; Guo, Z.H.; Zhang, Z.Y.; Han, C.G.; Wang, Y. Development of Beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol. J. 2019, 17, 1302–1315. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 2014, 9, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Marton, I.; Zuker, A.; Shklarman, E.; Zeevi, V.; Tovkach, A.; Roffe, S.; Ovadis, M.; Tzfira, T.; Vainstein, A. Nontransgenic genome modification in plant cells. Plant Physiol. 2010, 154, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Honig, A.; Marton, I.; Rosenthal, M.; Smith, J.J.; Nicholson, M.G.; Jantz, D.; Zuker, A.; Vainstein, A. Transient expression of virally delivered meganuclease in planta generates inherited genomic deletions. Mol. Plant. 2015, 8, 1292–1294. [Google Scholar] [CrossRef]
- Ali, Z.; Eid, A.; Ali, S.; Mahfouz, M.M. Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 2018, 244, 333–337. [Google Scholar] [CrossRef]
- Hu, J.; Li, S.; Li, Z.; Li, H.; Song, W.; Zhao, H.; Lai, J.; Xia, L.; Li, D.; Zhang, Y. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol. Plant Pathol. 2019, 20, 1463–1474. [Google Scholar] [CrossRef]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef]
- Ellison, E.E.; Nagalakshmi, U.; Gamo, M.E.; Huang, P.-j.; Dinesh-Kumar, S.; Voytas, D.F. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 2020, 6, 620–624. [Google Scholar] [CrossRef]
- Wu, M.; Wei, H.; Tan, H.; Pan, S.; Liu, Q.; Bejarano, E.R.; Lozano-Durán, R. Plant DNA polymerases α and δ mediate replication of geminiviruses. Nat. Commun. 2021, 12, 2780. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J.-k. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Aragonés, V.; Selma, S.; Vázquez-Vilar, M.; Orzáez, D.; Daròs, J.A. Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J. 2021, 106, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Ariga, H.; Toki, S.; Ishibashi, K. Potato virus X vector-mediated DNA-free genome editing in plants. Plant Cell Physiol. 2020, 61, 1946–1953. [Google Scholar] [CrossRef]
- Zhang, X.; Kang, L.; Zhang, Q.; Meng, Q.; Pan, Y.; Yu, Z.; Shi, N.; Jackson, S.; Zhang, X.; Wang, H. An RNAi suppressor activates in planta virus–mediated gene editing. Funct. Integr. Genom. 2020, 20, 471–477. [Google Scholar] [CrossRef]
- Kaya, H.; Ishibashi, K.; Toki, S. A split Staphylococcus aureus Cas9 as a compact genome-editing tool in plants. Plant Cell Physiol. 2017, 58, 643–649. [Google Scholar] [CrossRef]
- Gao, Q.; Xu, W.Y.; Yan, T.; Fang, X.D.; Cao, Q.; Zhang, Z.J.; Ding, Z.H.; Wang, Y.; Wang, X.B. Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 2019, 223, 2120–2133. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Sudarshana, M.; Jiang, H.; Rojas, M.R.; Lucas, W.J. Limitations on geminivirus genome size imposed by plasmodesmata and virus-encoded movement protein: Insights into DNA trafficking. Plant Cell 2003, 15, 2578–2591. [Google Scholar] [CrossRef]
- Regnard, G.L.; Halley-Stott, R.P.; Tanzer, F.L.; Hitzeroth, I.I.; Rybicki, E.P. High level protein expression in plants through the use of a novel autonomously replicating geminivirus shuttle vector. Plant Biotechnol. J. 2010, 8, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000, 19, 792–799. [Google Scholar] [CrossRef] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmood, M.A.; Naqvi, R.Z.; Rahman, S.U.; Amin, I.; Mansoor, S. Plant Virus-Derived Vectors for Plant Genome Engineering. Viruses 2023, 15, 531. https://doi.org/10.3390/v15020531
Mahmood MA, Naqvi RZ, Rahman SU, Amin I, Mansoor S. Plant Virus-Derived Vectors for Plant Genome Engineering. Viruses. 2023; 15(2):531. https://doi.org/10.3390/v15020531
Chicago/Turabian StyleMahmood, Muhammad Arslan, Rubab Zahra Naqvi, Saleem Ur Rahman, Imran Amin, and Shahid Mansoor. 2023. "Plant Virus-Derived Vectors for Plant Genome Engineering" Viruses 15, no. 2: 531. https://doi.org/10.3390/v15020531





