RBD and Spike DNA-Based Immunization in Rabbits Elicited IgG Avidity Maturation and High Neutralizing Antibody Responses against SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Ethics Statements
2.2. Production of RBD and Full-Length Spike SARS-CoV-2 Gene Plasmids
2.3. DNA-Based Immunization by Needle Injection and Serum Samples
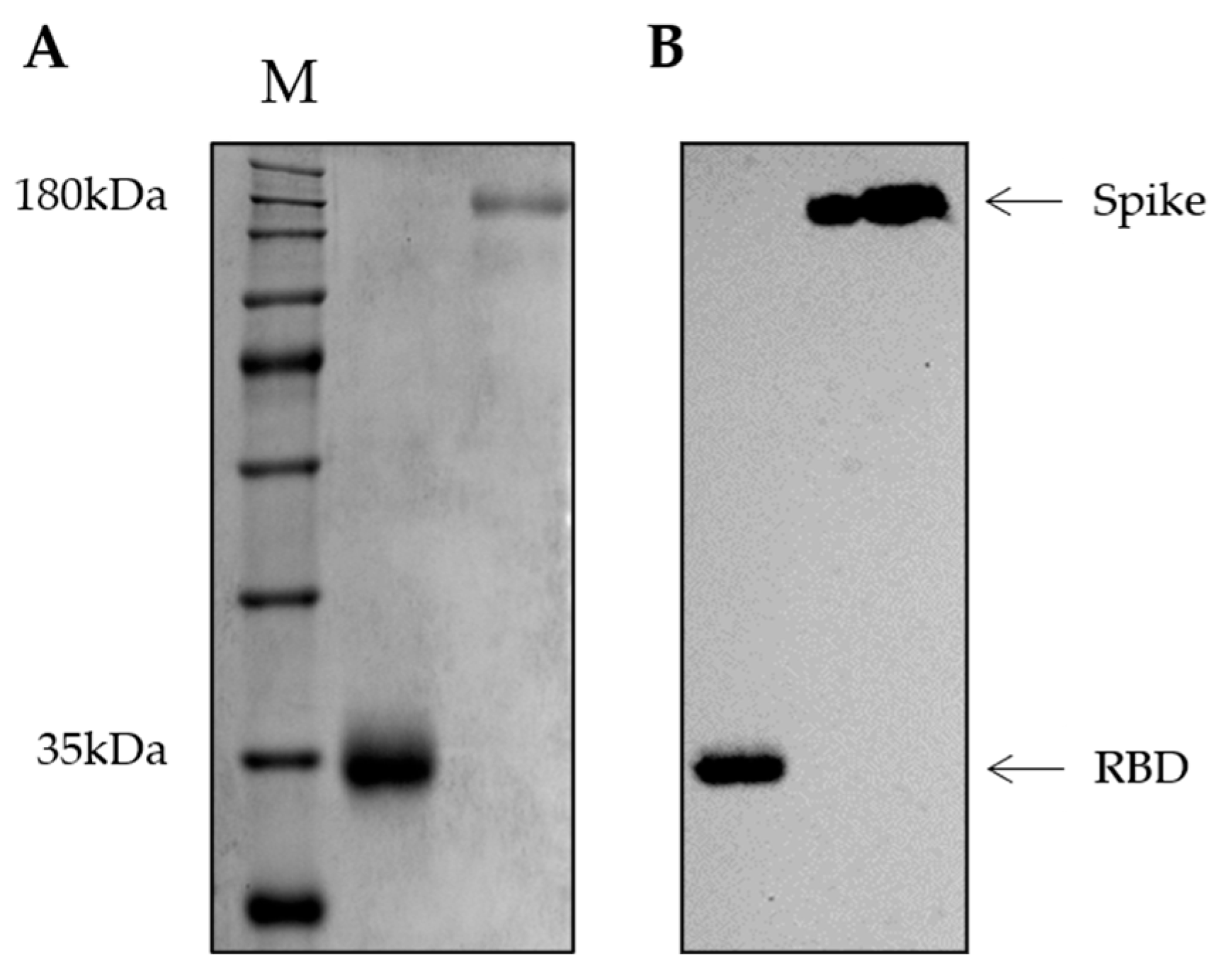
2.4. Production of RBD and Spike Recombinant Proteins
2.5. Immunoblotting Assay
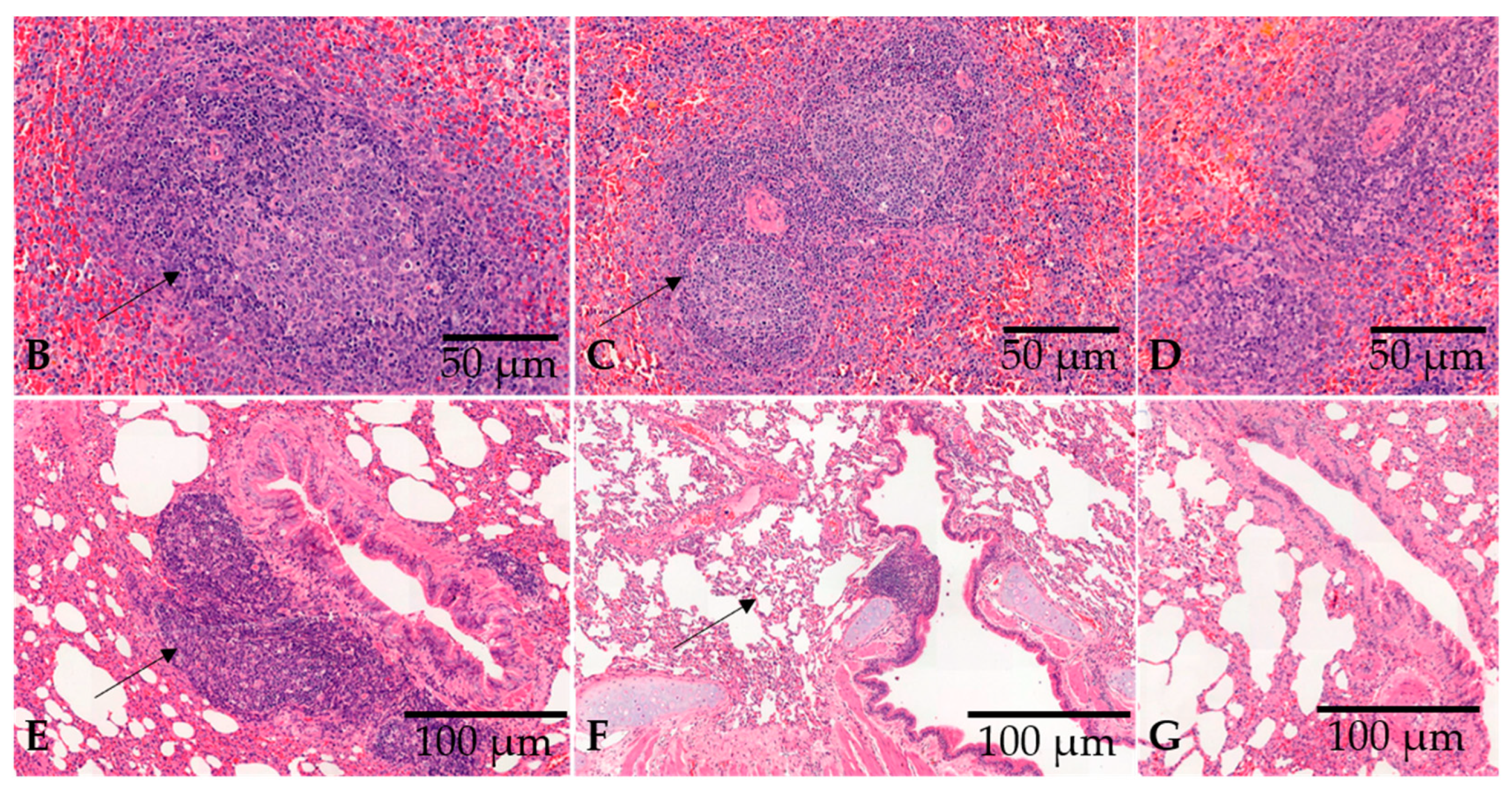
2.6. Pathological Examination and Histochemical Staining
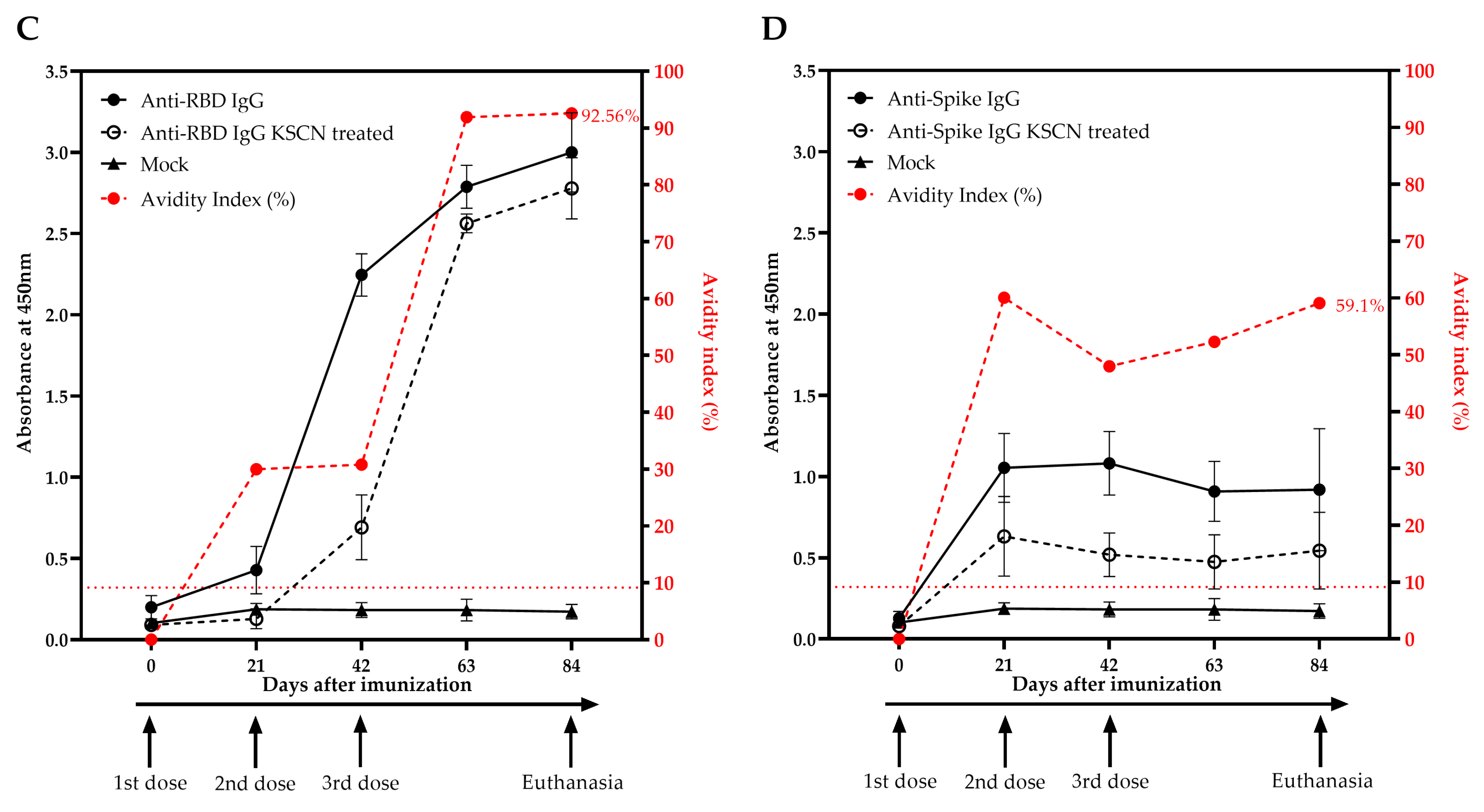
2.7. Indirect ELISA for Anti-RBD and Anti-Spike Specific IgG Antibodies Detection and Avidity Maturation Evaluation
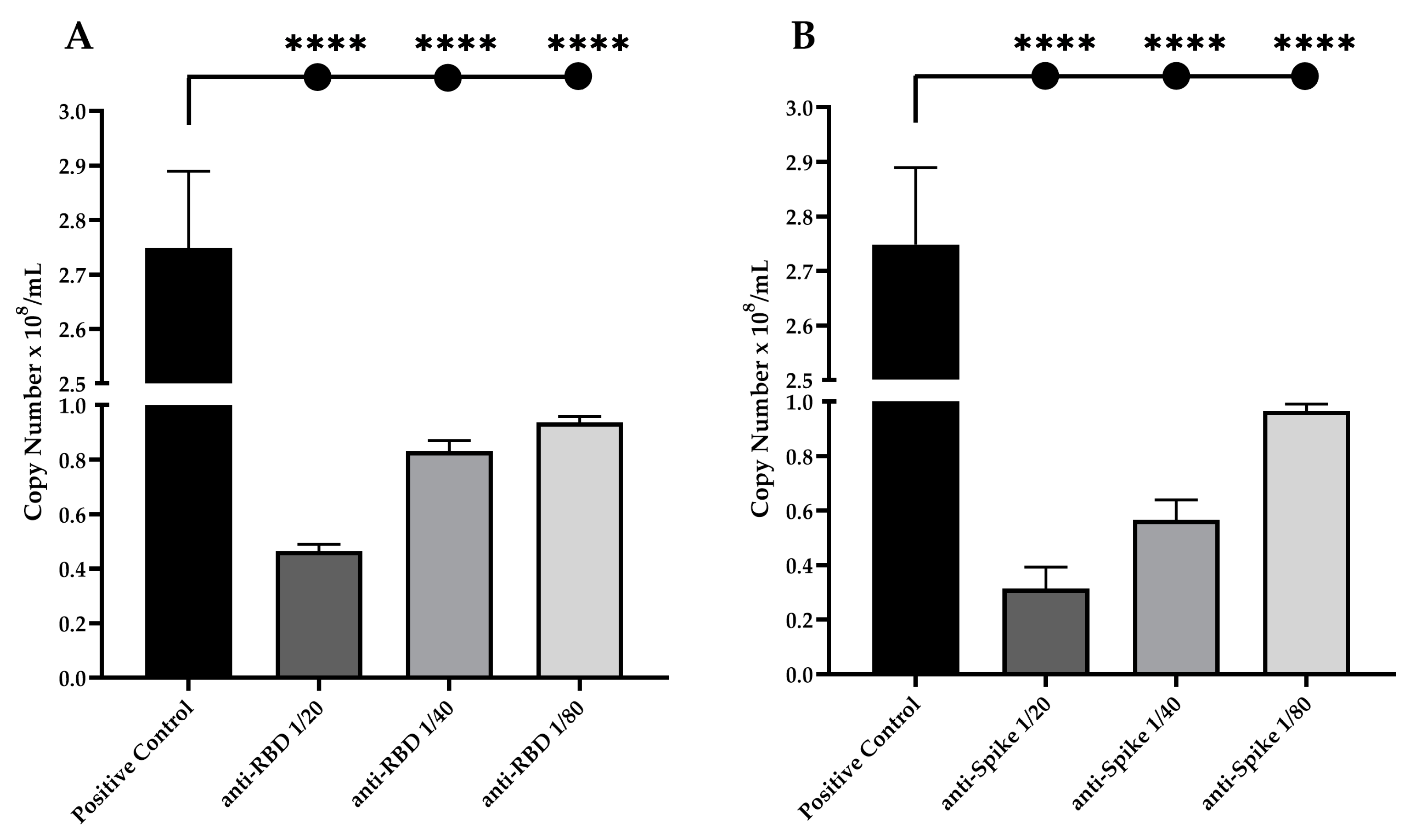
2.8. Neutralization Assay against Live SARS-CoV-2 Viruses
2.9. SARS-CoV-2 Pseudotyped Virus Neutralization Assays
2.10. Statistical and Data Analysis
3. Results
3.1. Plasmid Purification and Antigen Production
3.2. Vaccination with a DNA Plasmid by Needle Injection Elicits Robust Lymphoid Proliferation in the Spleen and Lung-Associated Tissues and High Titers of Anti-RBD and Anti-Spike IgG in Rabbits
3.3. Vaccination with a DNA Plasmid Elicits High Titers of Anti-RBD and Anti-Spike IgG in Rabbits
3.4. Neutralization Potential of Anti-RBD and Anti-Spike IgG Antibodies against Live and Pseudotyped SARS-CoV-2 Particles
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus That Causes a Global Threat. Int. J. Biol. Sci. 2020, 16, 1678. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.J.; Dong, X.; Cao, Y.Y.; Yuan, Y.D.; Yang, Y.B.; Yan, Y.Q.; Akdis, C.A.; Gao, Y.D. Clinical Characteristics of 140 Patients Infected with SARS-CoV-2 in Wuhan, China. Allergy Eur. J. Allergy Clin. Immunol. 2020, 75, 1730–1741. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadlbauer, D.; Amanat, F.; Chromikova, V.; Jiang, K.; Strohmeier, S.; Arunkumar, G.A.; Tan, J.; Bhavsar, D.; Capuano, C.; Kirkpatrick, E.; et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020, 57, e100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, E.B.; Prudencio, C.R.; De Gaspari, E. Experimental Studies Using OMV in a New Platform of SARS-CoV-2 Vaccines. Hum. Vaccines Immunother. 2021, 17, 2965–2968. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef]
- Li, Y.; Bi, Y.; Xiao, H.; Yao, Y.; Liu, X.; Hu, Z.; Duan, J.; Yang, Y.; Li, Z.; Li, Y.; et al. A Novel DNA and Protein Combination COVID-19 Vaccine Formulation Provides Full Protection against SARS-CoV-2 in Rhesus Macaques. Emerg. Microbes Infect. 2021, 10, 342–355. [Google Scholar] [CrossRef]
- Moura, A.D.; da Costa, H.H.M.; Correa, V.A.; Ana, A.K.; Lindoso, J.A.L.; De Gaspari, E.; Hong, M.A.; Cunha-Junior, J.P.; Prudencio, C.R. Assessment of Avidity Related to IgG Subclasses in SARS-CoV-2 Brazilian Infected Patients. Sci. Rep. 2021, 11, 17642. [Google Scholar] [CrossRef]
- Li, J.; Liu, Q.; Liu, J.; Fang, Z.; Luo, L.; Li, S.; Lei, Y.; Li, Z.; Jin, J.; Xie, R.; et al. Development of Bivalent MRNA Vaccines against SARS-CoV-2 Variants. Vaccines 2022, 10, 1807. [Google Scholar] [CrossRef]
- Takashita, E.; Yamayoshi, S.; Simon, V.; van Bakel, H.; Sordillo, E.M.; Pekosz, A.; Fukushi, S.; Suzuki, T.; Maeda, K.; Halfmann, P.; et al. Efficacy of Antibodies and Antiviral Drugs against Omicron BA.2.12.1, BA.4, and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 468–470. [Google Scholar] [CrossRef]
- WHO. Enhancing Response to Omicron SARS-CoV-2 Variant: Technical Brief and Priority Actions for Member States. Available online: https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states (accessed on 19 September 2022).
- Santos, F.R.D.S.; de Azevedo, M.S.P.; Bielavsky, M.; da Costa, H.H.M.; Ribeiro, D.G.; Nascimento, G.G.D.; Marcondes, G.M.P.; de Castro, B.P.; de Lima Neto, D.F.; Prudencio, C.R. Mutational Profile Confers Increased Stability of SARS-CoV-2 Spike Protein in Brazilian Isolates. J. Biomol. Struct. Dyn. 2021, 40, 13184–13189. [Google Scholar] [CrossRef] [PubMed]
- Astuti, I. Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lu, Y.; Xia, L.; Yuan, X.; Li, G.; Li, X.; Liu, L.; Liu, W.; Zhou, P.; Wang, C.Y.; et al. Analysis of 2019 Novel Coronavirus Infection and Clinical Characteristics of Outpatients: An Epidemiological Study from a Fever Clinic in Wuhan, China. J. Med. Virol. 2020, 92, 2758–2767. [Google Scholar] [CrossRef]
- Yang, H.S.; Racine-Brzostek, S.E.; Lee, W.T.; Hunt, D.; Yee, J.; Chen, Z.; Kubiak, J.; Cantu, M.; Hatem, L.; Zhong, E.; et al. SARS-CoV-2 Antibody Characterization in Emergency Department, Hospitalized and Convalescent Patients by Two Semi-Quantitative Immunoassays. Clin. Chim. Acta 2020, 509, 117–125. [Google Scholar] [CrossRef]
- Porter, K.R.; Raviprakash, K. DNA Vaccine Delivery and Improved Immunogenicity. Curr. Issues Mol. Biol. 2017, 22, 129–138. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent Advances in MRNA Vaccine Technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef]
- Pardi, N.; Muramatsu, H.; Weissman, D.; Karikó, K. In Vitro Transcription of Long RNA Containing. Methods Mol. Biol. 2013, 969, 29–42. [Google Scholar] [CrossRef]
- Wang, S.; Lu, S. DNA Immunization. Curr. Protoc. Microbiol. 2013, 4, 307–312. [Google Scholar] [CrossRef]
- Dey, A.; Rajanathan, T.M.C.; Chandra, H.; Pericherla, H.P.R.; Kumar, S.; Choonia, H.S.; Bajpai, M.; Singh, A.K.; Sinha, A.; Saini, G.; et al. Immunogenic Potential of DNA Vaccine Candidate, ZyCoV-D against SARS-CoV-2 in Animal Models. Vaccine 2021, 39, 4108–4116. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.A. DNA Vaccines: A Review. J. Intern. Med. 2003, 253, 402–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excler, J.L.; Saville, M.; Berkley, S.; Kim, J.H. Vaccine Development for Emerging Infectious Diseases. Nat. Med. 2021, 27, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ramos, S.J.; Bangalore, P.; Elwood, D.; Cashman, K.A.; Kudchodkar, S.B.; Schultheis, K.; Pugh, H.; Walters, J.; Tur, J.; et al. Multivalent DNA Vaccines as a Strategy to Combat Multiple Concurrent Epidemics: Mosquito-Borne and Hemorrhagic Fever Viruses. Viruses 2021, 13, 382. [Google Scholar] [CrossRef]
- Matić, Z.; Šantak, M. Current View on Novel Vaccine Technologies to Combat Human Infectious Diseases; Springer: Berlin/Heidelberg, Germany, 2022; Volume 106, ISBN 0123456789. [Google Scholar]
- Maslow, J.N. The Cost and Challenge of Vaccine Development for Emerging and Emergent Infectious Diseases. Lancet 2018, 6, e1266–e1267. [Google Scholar] [CrossRef] [Green Version]
- WHO, W.H.O. WHO Expert Committee on Biological Standardization, 56th ed.; World Health Organization: Geneva, Switzerland, 2007; ISBN 9789241209410. [Google Scholar]
- Silveira, M.M.; Moreira, G.M.S.G.; Mendonça, M. DNA Vaccines against COVID-19: Perspectives and Challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef]
- Bauer, G. High Avidity of Vaccine-Induced Immunoglobulin G against SARS-CoV-2: Potential Relevance for Protective Humoral Immunity. Explor. Immunol. 2022, 2, 133–156. [Google Scholar] [CrossRef]
- du Sert, N.P.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The Arrive Guidelines 2.0: Updated Guidelines for Reporting Animal Research. PLoS Biol. 2020, 18, 1769–1777. [Google Scholar] [CrossRef]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co. Ltd.: London, UK, 1959; ISBN 0900767782. [Google Scholar]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A Serological Assay to Detect SARS-CoV-2 Seroconversion in Humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef]
- Martins, P.; Machado, D.; Theizen, T.H.; Guarnieri, J.P.O.; Bernardes, B.G.; Gomide, G.P.; Corat, M.A.F.; Abbehausen, C.; Módena, J.L.P.; Melo, C.F.O.R.; et al. Outer Membrane Vesicles from Neisseria Meningitidis (Proteossome) Used for Nanostructured Zika Virus Vaccine Production. Sci. Rep. 2018, 8, 8290. [Google Scholar] [CrossRef]
- Corman, V.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Luisa Schmidt, M.; et al. Detection of 2019 -NCoV by RT-PCR. Euro Surveill 2020, 25, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, J.K.; Tang, T.; Nathan, L.; Jaimes, J.A.; Hsu, H.-L.; Daniel, S.; Whittaker, G.R. Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J. Vis. Exp. 2019, 145, 139–148. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Liu, B.; Wang, X.; Zhang, L. The Humoral Response and Antibodies against SARS-CoV-2 Infection. Nat. Immunol. 2022, 23, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Andreano, E.; Paciello, I.; Pierleoni, G.; Maccari, G.; Antonelli, G.; Abbiento, V.; Pileri, P.; Benincasa, L.; Giglioli, G.; Piccini, G.; et al. MRNA Vaccines and Hybrid Immunity Use Different B Cell Germlines to Neutralize Omicron BA.4 and BA.5. bioRxiv 2022. [Google Scholar] [CrossRef]
- He, X.; Hong, W.; Pan, X.; Lu, G.; Wei, X. SARS-CoV-2 Omicron Variant: Characteristics and Prevention. MedComm 2021, 2, 838–845. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Chan, P.K.S.; Lim, P.L.; Liu, E.Y.M.; Cheung, J.L.K.; Leung, D.T.M.; Sung, J.J.Y. Antibody Avidity Maturation during Severe Acute Respiratory Syndrome-Associated Coronavirus Infection. J. Infect. Dis. 2005, 192, 166–169. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, J.; Cui, X.; Zheng, J.; Li, R.; Wang, F.; Liu, J.; Hu, Y.-H. Evaluation of Immune Protection Induced by DNA Vaccines from Haemaphysalis Longicornis Paramyosin in Rabbits. Parasit. Vectors 2017, 10, 325. [Google Scholar] [CrossRef] [Green Version]
- Burgain, A.; Rochard, A.; Trollet, C.; Mazuet, C.; Popoff, M.R.; Escriou, V.; Scherman, D.; Bigey, P. DNA Electroporation in Rabbits as a Method for Generation of High-Titer Neutralizing Antisera Examples of the Botulinum Toxins Types A, B, and E. Hum. Vaccines Immunother. 2013, 9, 2147–2156. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhu, L.; Huang, W.; Tong, X.; Wu, H.; Tao, Y.; Tong, B.; Huang, H.; Chen, J.; Zhao, X.; et al. Potent RBD-Specific Neutralizing Rabbit Monoclonal Antibodies Recognize Emerging SARS- CoV-2 Variants Elicited by DNA Prime-Protein Boost Vaccination Emerging SARS-CoV-2 Variants Elicited by DNA Prime-Protein Boost. Emerg. Microbes Infect. 2021, 10, 1390–1403. [Google Scholar] [CrossRef]
- Ding, C.; Patel, D.; Ma, Y.; Mann, J.F.S.; Wu, J.; Gao, Y. Employing Broadly Neutralizing Antibodies as a Human Immunodeficiency Virus Prophylactic & Therapeutic Application. Front. Immunol. 2021, 12, 697683. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Wang, Q.; Wu, Q.; Chen, J.; Wang, X.; Wang, Y.; Chen, Y.; Xia, N. Rabbit Monoclonal Antibody Specifically Recognizing a Linear Epitope in the Rbd of Sars-Cov-2 Spike Protein. Vaccines 2021, 9, 829. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Coyle, E.M.; Klenow, L.; Tang, J.; Grubbs, G.; Liu, S.; Wang, T.; Golding, H.; Khurana, S. Antibody Signature Induced by SARS-CoV-2 Spike Protein Immunogens in Rabbits. Sci. Transl. Med. 2020, 12, eabc3539. [Google Scholar] [CrossRef] [PubMed]
- Verkhivker, G.; Agajanian, S.; Oztas, D.; Gupta, G. Dynamic Profiling of Binding and Allosteric Propensities of the SARS-CoV-2 Spike Protein with Different Classes of Antibodies: Mutational and Perturbation-Based Scanning Reveals the Allosteric Duality of Functionally Adaptable Hotspots. J. Chem. Theory Comput. 2021, 17, 4578–4598. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Henderson, R.; Edwards, R.J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S.M.; Kopp, M.; Li, D.; Parks, R.; Hsu, A.L.; et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020, 27, 925–933. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Sun, Z.; Zhu, X.; Marti, M.M.; Srivastava, S.S.; Berezuk, A.M.; Zhou, S.; Tuttle, K.S.; Sobolewski, M.D.; et al. SARS-CoV-2 Variants of Concern: Spike Protein Mutational Analysis and Epitope for Broad Neutralization. Nat. Commun. 2022, 13, 4696. [Google Scholar] [CrossRef]
- Díaz-Salinas, M.A.; Li, Q.; Ejemel, M.; Yurkovetskiy, L.; Luban, J.; Shen, K.; Wang, Y.; Munro, J.B. Conformational Dynamics and Allosteric Modulation of the SARS-CoV-2 Spike. eLife 2022, 11, e75433. [Google Scholar] [CrossRef]
- Anasir, M.I.; Poh, C.L. Structural Vaccinology for Viral Vaccine Design. Front. Microbiol. 2019, 10, 738. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-Terminal Domain Antigenic Mapping Reveals a Site of Vulnerability for SARS-CoV-2. Cell 2020, 184, 2332–2347. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A Neutralizing Human Antibody Binds to the N-Terminal Domain of the Spike Protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lu, S.; Jia, H.; Deng, Y.; Zhou, J.; Huang, B.; Yu, Y.; Lan, J.; Wang, W.; Lou, Y.; et al. A Novel Neutralizing Monoclonal Antibody Targeting the N-Terminal Domain of the MERS-CoV Spike Protein. Emerg. Microbes Infect. 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.E.; Park, Y.-J.; Stewart, C.; Brown, J.T.; Sharkey, W.K.; Walls, A.C.; Joshi, A.; Sprouse, K.R.; McCallum, M.; Tortorici, M.A.; et al. SARS-CoV-2 Spike Conformation Determines Plasma Neutralizing Activity Elicited by a Wide Panel of Human Vaccines. Sci. Immunol. 2022, 7, eadf1421. [Google Scholar] [CrossRef] [PubMed]
- Williams, J. Vector Design for Improved DNA Vaccine Efficacy, Safety and Production. Vaccines 2013, 1, 225–249. [Google Scholar] [CrossRef]
- Oliveira, S.C.; Rosìnha, G.M.S.; De-Brito, C.F.A.; Fonseca, C.T.; Afonso, R.R.; Costa, M.C.M.S.; Goes, A.M.; Rech, E.L.; Azevedo, V. Immunological Properties of Gene Vaccines Delivered by Different Routes. Brazilian J. Med. Biol. Res. 1999, 32, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Bauer, G. The Potential Significance of High Avidity Immunoglobulin G (IgG) for Protective Immunity towards SARS-CoV-2. Int. J. Infect. Dis. 2021, 106, 61–64. [Google Scholar] [CrossRef]
- Struck, F.; Schreiner, P.; Staschik, E.; Wochinz-Richter, K.; Schulz, S.; Soutschek, E.; Motz, M.; Bauer, G. Vaccination versus Infection with SARS-CoV-2: Establishment of a High Avidity IgG Response versus Incomplete Avidity Maturation. J. Med. Virol. 2021, 93, 6765–6777. [Google Scholar] [CrossRef]
- Iwasaki, A.; Yang, Y. The Potential Danger of Suboptimal Antibody Responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 339–341. [Google Scholar] [CrossRef] [Green Version]
- Puschnik, A.; Lau, L.; Cromwell, E.A.; Balmaseda, A.; Zompi, S.; Harris, E. Correlation between Dengue-Specific Neutralizing Antibodies and Serum Avidity in Primary and Secondary Dengue Virus 3 Natural Infections in Humans. PLoS Negl. Trop. Dis. 2013, 7, e2274. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, E.B.; De Gaspari, E. Avidity Assay to Test Functionality of Anti-SARS-CoV-2 Antibodies. Vaccine 2021, 39, 1473–1475. [Google Scholar] [CrossRef]
- Zhou, R.; To, K.K.-W.; Wong, Y.-C.; Hung, I.F.-N.; Yuen, K.-Y.; Chen, Z. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877. [Google Scholar] [CrossRef] [PubMed]
- Khatri, I.; Staal, F.J.T.; van Dongen, J.J.M. Blocking of the High-Affinity Interaction-Synapse Between SARS-CoV-2 Spike and Human ACE2 Proteins Likely Requires Multiple High-Affinity Antibodies: An Immune Perspective. Front. Immunol. 2020, 11, 570018. [Google Scholar] [CrossRef] [PubMed]
- Correa, V.A.; Rodrigues, T.S.; Portilho, A.I.; Trzewikoswki de Lima, G.; De Gaspari, E. Modified ELISA for Antibody Avidity Evaluation: The Need for Standardization. Biomed. J. 2021, 44, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Subbarao, N. Insilico Study on the Effect of SARS-CoV-2 RBD Hotspot Mutants’ Interaction with ACE2 to Understand the Binding Affinity and Stability. Virology 2020, 561, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Nelson, G.; Olson, C.A.; Buzko, O.; Higashide, W.; Shin, A.; Gonzalez, M.; Taft, J.; Patel, R.; Buta, S.; et al. An ACE2 Triple Decoy That Neutralizes SARS-CoV-2 Shows Enhanced Affinity for Virus Variants. Sci. Rep. 2021, 11, 12740. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Liu, Y.; Lei, Z.; Dicker, J.; Cao, Y.; Zhang, X.F.; Im, W. Differential Interactions between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. J. Chem. Theory Comput. 2021, 17, 7972–7979. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, H.H.M.; Orts, D.J.B.; Moura, A.D.; Duarte-Neto, A.N.; Cirqueira, C.S.; Réssio, R.A.; Kanamura, C.T.; Miguita, K.; Ferreira, J.E.; Santos, R.T.M.; et al. RBD and Spike DNA-Based Immunization in Rabbits Elicited IgG Avidity Maturation and High Neutralizing Antibody Responses against SARS-CoV-2. Viruses 2023, 15, 555. https://doi.org/10.3390/v15020555
da Costa HHM, Orts DJB, Moura AD, Duarte-Neto AN, Cirqueira CS, Réssio RA, Kanamura CT, Miguita K, Ferreira JE, Santos RTM, et al. RBD and Spike DNA-Based Immunization in Rabbits Elicited IgG Avidity Maturation and High Neutralizing Antibody Responses against SARS-CoV-2. Viruses. 2023; 15(2):555. https://doi.org/10.3390/v15020555
Chicago/Turabian Styleda Costa, Hernan H. M., Diego J. B. Orts, Andrew D. Moura, Amaro N. Duarte-Neto, Cinthya S. Cirqueira, Rodrigo A. Réssio, Cristina T. Kanamura, Karen Miguita, Jerenice E. Ferreira, Raimunda T. M. Santos, and et al. 2023. "RBD and Spike DNA-Based Immunization in Rabbits Elicited IgG Avidity Maturation and High Neutralizing Antibody Responses against SARS-CoV-2" Viruses 15, no. 2: 555. https://doi.org/10.3390/v15020555







