Comparison of Pan-Lyssavirus RT-PCRs and Development of an Improved Protocol for Surveillance of Non-RABV Lyssaviruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Silico Evaluation
2.2. Improvement of LN34 rRT-PCR Assay
2.3. Sample Panel Selection
2.4. Comparison of Lyssavirus Detection Methods
2.5. Repeatability and Analytical Specificity (ASp) of (n) LN34
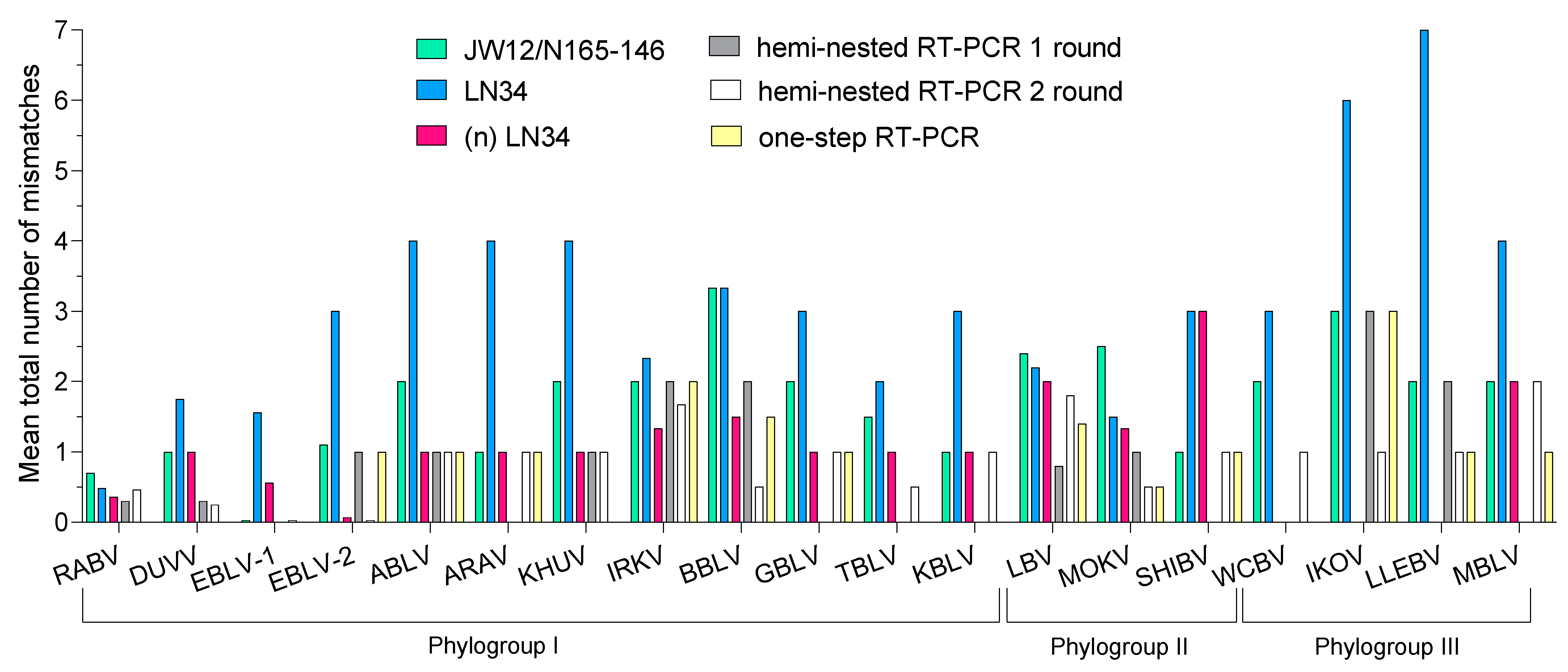
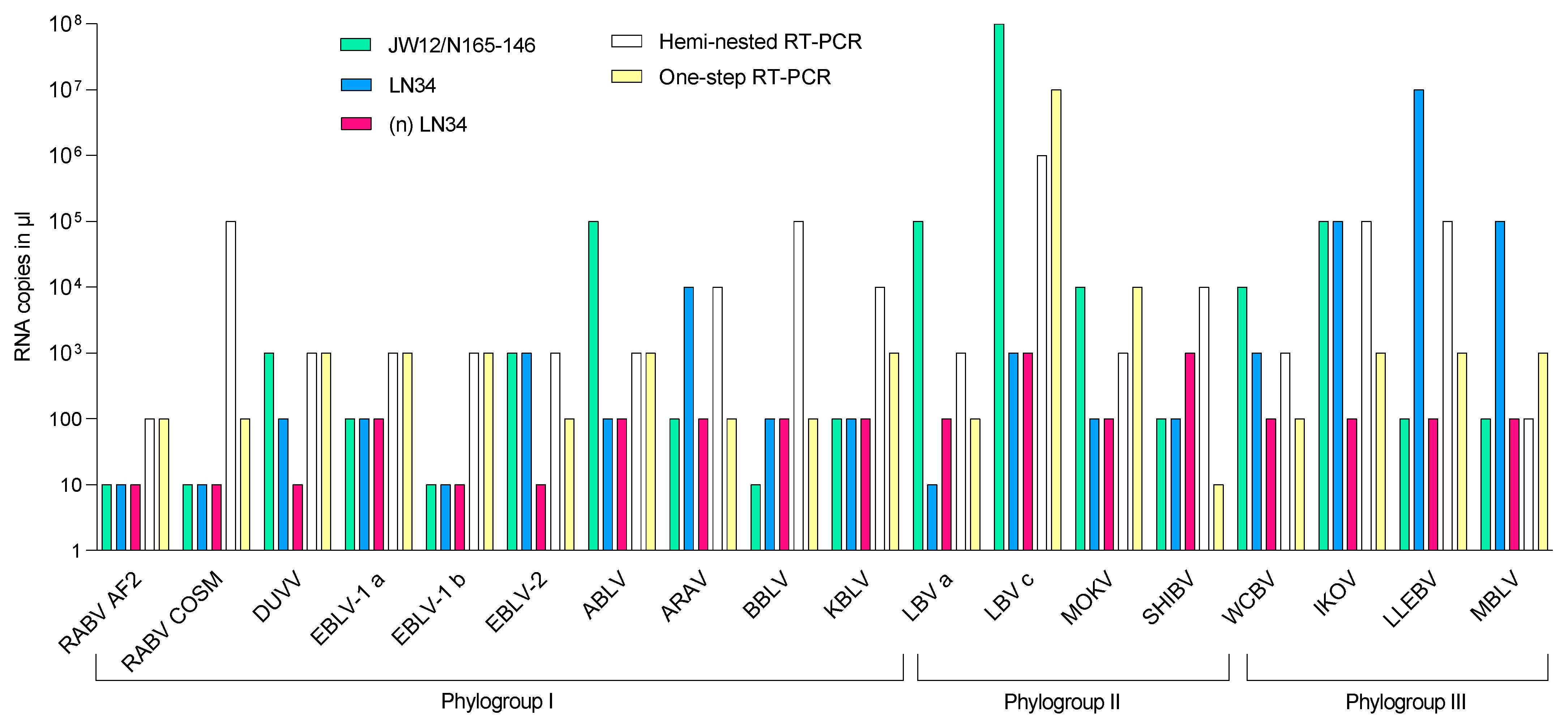
3. Results
3.1. In Silico Evaluation
3.2. (n) LN34 Assay Development
3.3. Analytical Sensitivity (ASe)
3.4. (n) LN34 Specificity and Repeatability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fooks, A.R.; Cliquet, F.; Finke, S.; Freuling, C.; Hemachudha, T.; Mani, R.S.; Müller, T.; Nadin-Davis, S.; Picard-Meyer, E.; Wilde, H.; et al. Rabies. Nat. Rev. Dis. Prim. 2017, 3, 17091. [Google Scholar] [CrossRef]
- World Health Organization; Food and Agriculture Organization of the United Nations; World Organisation for Animal Health; Global Alliance for Rabies Control. Zero by 30: The Global Strategic Plan to End Human Deaths from Dog-Mediated Rabies by 2030: United Against Rabies Collaboration: First Annual Progress Report: Global Strategic Plan to End Human Deaths from Dog-Mediated Rabies by 2030; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Marston, D.A.; Banyard, A.C.; McElhinney, L.M.; Freuling, C.M.; Finke, S.; de Lamballerie, X.; Müller, T.; Fooks, A.R. The Lyssavirus Host-Specificity Conundrum—Rabies Virus—The Exception Not the Rule. Curr. Opin. Virol. 2018, 28, 68–73. [Google Scholar] [CrossRef]
- Velasco-Villa, A.; Mauldin, M.R.; Shi, M.; Escobar, L.E.; Gallardo-Romero, N.F.; Damon, I.; Olson, V.A.; Streicker, D.G.; Emerson, G. The History of Rabies in the Western Hemisphere. Antiviral Res. 2017, 146, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Genus: Lyssavirus—Rhabdoviridae—Negative-Sense RNA Viruses—ICTV. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/rhabdoviridae/795/genus-lyssavirus (accessed on 4 August 2022).
- Coertse, J.; Grobler, C.S.; Sabeta, C.T.; Seamark, E.C.J.; Kearney, T.; Paweska, J.T.; Markotter, W. Lyssaviruses in Insectivorous Bats, South Africa, 2003–2018. Emerg Infect. Dis. 2020, 26, 3056–3060. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (Ed.) WHO Expert Consultation on Rabies: Third Report; WHO technical report series; World Health Organization: Geneva, Switzerland, 2018; ISBN 978-92-4-121021-8. [Google Scholar]
- Fooks, A.R.; Shipley, R.; Markotter, W.; Tordo, N.; Freuling, C.M.; Müller, T.; McElhinney, L.M.; Banyard, A.C.; Rupprecht, C.E. Renewed Public Health Threat from Emerging Lyssaviruses. Viruses 2021, 13, 1769. [Google Scholar] [CrossRef] [PubMed]
- Calvelage, S.; Tammiranta, N.; Nokireki, T.; Gadd, T.; Eggerbauer, E.; Zaeck, L.M.; Potratz, M.; Wylezich, C.; Höper, D.; Müller, T.; et al. Genetic and Antigenetic Characterization of the Novel Kotalahti Bat Lyssavirus (KBLV). Viruses 2021, 13, 69. [Google Scholar] [CrossRef]
- Leopardi, S.; Barneschi, E.; Manna, G.; Zecchin, B.; Priori, P.; Drzewnioková, P.; Festa, F.; Lombardo, A.; Parca, F.; Scaravelli, D.; et al. Spillover of West Caucasian Bat Lyssavirus (WCBV) in a Domestic Cat and Westward Expansion in the Palearctic Region. Viruses 2021, 13, 2064. [Google Scholar] [CrossRef]
- Murray, K.A.; Morgan, J.; Allworth, A. A Human Case of Encephalitis Due to a Lyssavirus Recently Identified in Fruit Bats. Commun. Dis. Intell. 1996, 20, 504. [Google Scholar]
- van Thiel, P.-P.A.M.; de Bie, R.M.A.; Eftimov, F.; Tepaske, R.; Zaaijer, H.L.; van Doornum, G.J.J.; Schutten, M.; Osterhaus, A.D.M.E.; Majoie, C.B.L.M.; Aronica, E.; et al. Fatal Human Rabies Due to Duvenhage Virus from a Bat in Kenya: Failure of Treatment with Coma-Induction, Ketamine, and Antiviral Drugs. PLOS Negl. Trop. Dis. 2009, 3, e428. [Google Scholar] [CrossRef] [Green Version]
- Nathwani, D.; McIntyre, P.G.; White, K.; Shearer, A.J.; Reynolds, N.; Walker, D.; Orange, G.V.; Fooks, A.R. Fatal Human Rabies Caused by European Bat Lyssavirus Type 2a Infection in Scotland. Clin. Infect. Dis. 2003, 37, 598–601. [Google Scholar] [CrossRef]
- Regnault, B.; Evrard, B.; Plu, I.; Dacheux, L.; Troadec, E.; Cozette, P.; Chrétien, D.; Duchesne, M.; Vallat, J.-M.; Jamet, A.; et al. First Case of Lethal Encephalitis in Western Europe Due to European Bat Lyssavirus Type 1. Clin. Infect. Dis. 2022, 74, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Méthodes|EURL. Available online: https://eurl-rabies.anses.fr/en/minisite/rabies/methods (accessed on 28 July 2022).
- OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2018. OIE—World Organisation for Animal Health. Available online: http://www.oie.int/standard-setting/terrestrial-manual/access-online/ (accessed on 11 February 2019).
- Robardet, E.; Picard-Meyer, E.; Andrieu, S.; Servat, A.; Cliquet, F. International Interlaboratory Trials on Rabies Diagnosis: An Overview of Results and Variation in Reference Diagnosis Techniques (Fluorescent Antibody Test, Rabies Tissue Culture Infection Test, Mouse Inoculation Test) and Molecular Biology Techniques. J. Virol. Methods 2011, 177, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, C.E.; Fooks, A.R.; Abela-Ridder, B. (Eds.) Laboratory Techniques in Rabies—Volume 2, 5th ed.; World Health Organization: Geneva, Switzerland, 2018; ISBN 978-92-4-151530-6. [Google Scholar]
- OIE Principles and Methods of Validation of Diagnostic Assays for Infectious Diseases. Available online: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/1.01.06_VALIDATION.pdf (accessed on 11 February 2019).
- Robardet, E.; Servat, A.; Rieder, J.; Picard-Meyer, E.; Cliquet, F. Multi-Annual Performance Evaluation of Laboratories in Post-Mortem Diagnosis of Animal Rabies: Which Techniques Lead to the Most Reliable Results in Practice? PLoS Negl. Trop. Dis. 2021, 15, e0009111. [Google Scholar] [CrossRef]
- Robardet, E.; Andrieu, S.; Rasmussen, T.B.; Dobrostana, M.; Horton, D.L.; Hostnik, P.; Jaceviciene, I.; Juhasz, T.; Müller, T.; Mutinelli, F.; et al. Comparative Assay of Fluorescent Antibody Test Results among Twelve European National Reference Laboratories Using Various Anti-Rabies Conjugates. J. Virol. Methods 2013, 191, 88–94. [Google Scholar] [CrossRef]
- Banyard, A.C.; Selden, D.; Wu, G.; Thorne, L.; Jennings, D.; Marston, D.; Finke, S.; Freuling, C.M.; Müller, T.; Echevarría, J.E.; et al. Isolation, Antigenicity and Immunogenicity of Lleida Bat Lyssavirus. J. Gen. Virol. 2018, 99, 1590–1599. [Google Scholar] [CrossRef]
- Heaton, P.R.; Johnstone, P.; McElhinney, L.M.; Cowley, R.; O’Sullivan, E.; Whitby, J.E. Heminested PCR Assay for Detection of Six Genotypes of Rabies and Rabies-Related Viruses. J. Clin. Microbiol. 1997, 35, 2762–2766. [Google Scholar] [CrossRef] [Green Version]
- Wakeley, P.R.; Johnson, N.; McElhinney, L.M.; Marston, D.; Sawyer, J.; Fooks, A.R. Development of a Real-Time, TaqMan Reverse Transcription-PCR Assay for Detection and Differentiation of Lyssavirus Genotypes 1, 5, and 6. J. Clin. Microbiol. 2005, 43, 2786–2792. [Google Scholar] [CrossRef] [Green Version]
- De Benedictis, P.; De Battisti, C.; Dacheux, L.; Marciano, S.; Ormelli, S.; Salomoni, A.; Caenazzo, S.T.; Lepelletier, A.; Bourhy, H.; Capua, I.; et al. Lyssavirus Detection and Typing Using Pyrosequencing. J. Clin. Microbiol. 2011, 49, 1932–1938. [Google Scholar] [CrossRef] [Green Version]
- Gigante, C.M.; Dettinger, L.; Powell, J.W.; Seiders, M.; Condori, R.E.C.; Griesser, R.; Okogi, K.; Carlos, M.; Pesko, K.; Breckenridge, M.; et al. Multi-Site Evaluation of the LN34 Pan-Lyssavirus Real-Time RT-PCR Assay for Post-Mortem Rabies Diagnostics. PLoS ONE 2018, 13, e0197074. [Google Scholar] [CrossRef]
- Hoffmann, B.; Freuling, C.M.; Wakeley, P.R.; Rasmussen, T.B.; Leech, S.; Fooks, A.R.; Beer, M.; Müller, T. Improved Safety for Molecular Diagnosis of Classical Rabies Viruses by Use of a TaqMan Real-Time Reverse Transcription-PCR “Double Check” Strategy. J. Clin. Microbiol. 2010, 48, 3970–3978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caraballo, D.A.; Lombardo, M.A.; Becker, P.; Sabio, M.S.; Lema, C.; Martínez, L.M.; Beltrán, F.J.; Li, Y.; Cisterna, D.M. Evaluation of Two Real-Time, TaqMan Reverse Transcription-PCR Assays for Detection of Rabies Virus in Circulating Variants from Argentina: Influence of Sequence Variation. Viruses 2020, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Hayman, D.T.S.; Banyard, A.C.; Wakeley, P.R.; Harkess, G.; Marston, D.; Wood, J.L.N.; Cunningham, A.A.; Fooks, A.R. A Universal Real-Time Assay for the Detection of Lyssaviruses. J. Virol. Methods 2011, 177, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marston, D.A.; Jennings, D.L.; MacLaren, N.C.; Dorey-Robinson, D.; Fooks, A.R.; Banyard, A.C.; McElhinney, L.M. Pan-Lyssavirus Real Time RT-PCR for Rabies Diagnosis. J. Vis. Exp. 2019, 149, e59709. [Google Scholar] [CrossRef] [Green Version]
- Wadhwa, A.; Wilkins, K.; Gao, J.; Condori Condori, R.E.; Gigante, C.M.; Zhao, H.; Ma, X.; Ellison, J.A.; Greenberg, L.; Velasco-Villa, A.; et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR Assay for the Detection of Highly Variable Rabies Virus and Other Lyssaviruses. PLoS Negl. Trop. Dis. 2017, 11, e0005258. [Google Scholar] [CrossRef]
- Albertini, A.A.V.; Ruigrok, R.W.H.; Blondel, D. Chapter 1—Rabies Virus Transcription and Replication. In Advances in Virus Research; Jackson, A.C., Ed.; Research Advances in Rabies; Academic Press: Cambridge, MA, USA, 2011; Volume 79, pp. 1–22. [Google Scholar]
- Omodo, M.; Ar Gouilh, M.; Mwiine, F.N.; Okurut, A.R.A.; Nantima, N.; Namatovu, A.; Nakanjako, M.F.; Isingoma, E.; Arinaitwe, E.; Esau, M.; et al. Rabies in Uganda: Rabies Knowledge, Attitude and Practice and Molecular Characterization of Circulating Virus Strains. BMC Infect. Dis. 2020, 20, 200. [Google Scholar] [CrossRef]
- Traoré, A.; Picard-Meyer, E.; Mauti, S.; Biarnais, M.; Balmer, O.; Samaké, K.; Kamissoko, B.; Tembely, S.; Sery, A.; Traoré, A.K.; et al. Molecular Characterization of Canine Rabies Virus, Mali, 2006–2013. Emerg. Infect. Dis. 2016, 22, 866–870. [Google Scholar] [CrossRef] [Green Version]
- Mbuyi, G.T.; Kawaya, E.K.; Twabela, A.T.; Cattoli, G.; Walandila, J.S.; Naletoski, I.; Masumu, J.; Dundon, W.G. Molecular Characterization of Rabies Viruses from Two Western Provinces of the Democratic Republic of the Congo (2008–2017). Virus Genes 2020, 56, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Gourlaouen, M.; Angot, A.; Mancin, M.; Bebay, C.; Soumaré, B.; Ellero, F.; Zecchin, B.; Leopardi, S.; Battisti, C.D.; Terregino, C.; et al. An Inter-Laboratory Trial as a Tool to Increase Rabies Diagnostic Capabilities of Sub-Saharan African Veterinary Laboratories. PLoS Negl. Trop. Dis. 2020, 14, e0008010. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Freuling, C.M.; Müller, T.; Wegelt, A.; Kooi, E.A.; Rasmussen, T.B.; Voller, K.; Marston, D.A.; Fooks, A.R.; Beer, M.; et al. Molecular Double-Check Strategy for the Identification and Characterization of European Lyssaviruses. J. Virol. Methods 2014, 203, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Picard-Meyer, E.; Peytavin de Garam, C.; Schereffer, J.L.; Marchal, C.; Robardet, E.; Cliquet, F. Cross-Platform Evaluation of Commercial Real-Time SYBR Green RT-PCR Kits for Sensitive and Rapid Detection of European Bat Lyssavirus Type 1. BioMed Res. Int. 2015, 2015, 8395. [Google Scholar] [CrossRef] [Green Version]
- Conselheiro, J.; Barone, G.; Taniwaki, S.; Silva, S.; Agostinho, W.; Aguiar, J.; Brandão, P. Evolution of Rabies Virus Isolates: Virulence Signatures and Effects of Modulation by Neutralizing Antibodies. Pathogens 2022, 11, 1556. [Google Scholar] [CrossRef] [PubMed]
- Nitschel, S.; Zaeck, L.M.; Potratz, M.; Nolden, T.; te Kamp, V.; Franzke, K.; Höper, D.; Pfaff, F.; Finke, S. Point Mutations in the Glycoprotein Ectodomain of Field Rabies Viruses Mediate Cell Culture Adaptation through Improved Virus Release in a Host Cell Dependent and Independent Manner. Viruses 2021, 13, 1989. [Google Scholar] [CrossRef] [PubMed]
- Coertse, J.; Geldenhuys, M.; le Roux, K.; Markotter, W. Lagos Bat Virus, an Under-Reported Rabies-Related Lyssavirus. Viruses 2021, 13, 576. [Google Scholar] [CrossRef] [PubMed]
- Shipley, R.; Wright, E.; Selden, D.; Wu, G.; Aegerter, J.; Fooks, A.R.; Banyard, A.C. Bats and Viruses: Emergence of Novel Lyssaviruses and Association of Bats with Viral Zoonoses in the EU. Trop. Med. Infect. Dis. 2019, 4, 31. [Google Scholar] [CrossRef] [Green Version]
- Dacheux, L.; Larrous, F.; Mailles, A.; Boisseleau, D.; Delmas, O.; Biron, C.; Bouchier, C.; Capek, I.; Muller, M.; Ilari, F.; et al. European Bat Lyssavirus Transmission among Cats, Europe. Emerg. Infect. Dis. 2009, 15, 280–284. [Google Scholar] [CrossRef]
- Marston, D.; McElhinney, L.; Johnson, N.; Müller, T.; Conzelmann, K.-K.; Tordo, N.; Fooks, A.R. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G–L 3′ non-translated region. J. Gen. Virol. 2007, 88, 1302–1314. [Google Scholar] [CrossRef]
| Assay | Target | Name | Type | S/As | Sequence (5′ to 3′) |
|---|---|---|---|---|---|
| JW12 | primer | S | ATGTAACACCYCTACAATG | ||
| Hemi-nested [18,23] | N gene | JW6 UNI | primer | As | ARTTVGCRCACATYTTRTG |
| JW10 UNI | primer | As | GTCATYARWGTRTGRTGYTC | ||
| One-step [25] | N gene | RabForPyro | primer | S | AACACYYCTACAATGGA |
| RabRev1Pyro | primer | As | TCCAATTNGCACACATTTTGTG | ||
| RabRev2Pyro | primer | As | TCCARTTAGCGCACATYTTATG | ||
| RabRev3Pyro | primer | As | TCCAGTTGGCRCACATCTTRTG | ||
| JW12/N165-146 [18,24] | N gene | JW12 | primer | S | ATGTAACACCYCTACAATG |
| N165-145 | primer | As | GCAGGGTAYTTRTACTCATA | ||
| LN34 [18,26] | 3′UTR and N gene | LN34 FW1 | primer | S | ACGCTTAACAACCAGATCAAAGAA |
| LN34 FW2 | primer | S | ACGCTTAACAACAAAATCADAGAAG | ||
| LN34Probe | probe | S | FAM-AACACCYCTACAATGGA-BHQ1 | ||
| LN34lago Probe | probe | S | FAM-AACACTACTACAATGGA-BHQ1 | ||
| LN34REV | primer | As | CMGGGTAYTTRTAYTCATAYTGRTC |
| Phylogroup | Species | Virus Abbreviation | Virus Name/Clade or Lineage | Genbank Accession Number |
|---|---|---|---|---|
| I | Lyssavirus rabies | RABV AF2 | rabies virus/lineage Africa 2 | MK471246 |
| RABV COSM | rabies virus/lineage Cosmopolitan | KR906742 | ||
| Lyssavirus duvenhage | DUVV | Duvenhage virus | EU293120 | |
| Lyssavirus hamburg | EBLV-1 a | European bat lyssavirus type 1/lineage a | MF187809 | |
| EBLV-1 b | European bat lyssavirus type 1/lineage b | MF187859 | ||
| Lyssavirus helsinki | EBLV-2 | European bat lyssavirus type 2 | KY688150 | |
| Lyssavirus australis | ABLV | Australian bat lyssavirus | AY573937 | |
| Lyssavirus aravan | ARAV | Aravan virus | NC_020808 | |
| Lyssavirus bokeloh | BBLV | Bokeloh bat lyssavirus | NC_025251 | |
| unclassified | KBLV | Kotalahti bat lyssavirus | LR994545 | |
| II | Lyssavirus lagos | LBV a | Lagos bat virus/lineage a | EU293108 |
| LBV c | Lagos bat virus/lineage c | EF547449 | ||
| Lyssavirus mokola | MOKV | Mokola virus | EU293118 | |
| Lyssavirus shimoni | SHIBV | Shimoni bat virus | NC_025365 | |
| III | Lyssavirus caucasicus | WCBV | West Caucasian bat virus | KY688150 |
| Lyssavirus ikoma | IKOV | Ikoma lyssavirus | NC_018629 | |
| Lyssavirus lleida | LLBV | Lleida bat lyssavirus | MG983927 | |
| unclassified | MBLV | Matlo bat lyssavirus | MW653808 |
| TNMM * | rRT-PCR | RT-PCR | ||||
|---|---|---|---|---|---|---|
| LN34 | JW12/N165-146 | Hemi-Nested | One-Step | |||
| Primers | Probes | 1st Round | 2nd Round | |||
| Phylogroup I (n = 251) | ||||||
| 0 | 31% | 99% | 55% | 70% | 71% | 70% |
| 1 | 27% | 1% | 28% | 29% | 25% | 29% |
| 2 | 25% | 0% | 14% | 1% | 4% | 1% |
| 3 | 15% | 0% | 2% | 0% | 0% | 0% |
| 4 | 2% | 0% | 0% | 0% | 0% | 0% |
| 5 | 0% | 0% | 1% | 0% | 0% | 0% |
| 6 | 0% | 0% | 0% | 0% | 0% | 0% |
| 7 | 0% | 0% | 0% | 0% | 0% | 0% |
| AC ** | 83% | 100% | 97% | 100% | 100% | 100% |
| Phylogroup II (n = 12) | ||||||
| 0 | 0% | 50% | 8% | 8% | 25% | 25% |
| 1 | 33% | 50% | 17% | 83% | 67% | 58% |
| 2 | 42% | 0% | 25% | 0% | 8% | 0% |
| 3 | 25% | 0% | 42% | 8% | 0% | 17% |
| 4 | 0% | 0% | 0% | 0% | 0% | 0% |
| 5 | 0% | 0% | 8% | 0% | 0% | 0% |
| 6 | 0% | 0% | 0% | 0% | 0% | 0% |
| 7 | 0% | 0% | 0% | 0% | 0% | 0% |
| AC ** | 75% | 100% | 50% | 91% | 100% | 83% |
| Phylogroup III (n = 6) | ||||||
| 0 | 0% | 83% | 0% | 33% | 50% | 0% |
| 1 | 0% | 17% | 0% | 50% | 0% | 83% |
| 2 | 0% | 0% | 83% | 0% | 33% | 17% |
| 3 | 33% | 0% | 17% | 17% | 17% | 0% |
| 4 | 17% | 0% | 0% | 0% | 0% | 0% |
| 5 | 0% | 0% | 0% | 0% | 0% | 0% |
| 6 | 17% | 0% | 0% | 0% | 0% | 0% |
| 7 | 33% | 0% | 0% | 0% | 0% | 0% |
| AC ** | 0% | 100% | 83% | 83% | 83% | 100% |
| Name | S/As | Sequence (5′ to 3′) | Length (nt) | Tm (°C) |
|---|---|---|---|---|
| FW1 | S | ACGCTTAACAACMARATCAAAGAA | 24 | 56.6–59.2 |
| FW2 | S | ACGCTTAACAACAAAATCADARAAG | 25 | 55.7–58.6 |
| FW3 | S | ACGCTTAACGACAAAAHCAGARAAG | 25 | 59.1–62.0 |
| FW4 | S | ACGCTTAACAGCTAAAAACYAGAAG | 25 | 57.9–60.3 |
| FW5 | S | ACGCTTAACARCAAAATCTTATAAG | 25 | 54.7–56.5 |
| REV1 | As | Same as original (LN34 REV) [26] | 25 | 51.5–62.2 |
| REV2 | As | CTGGATATTTGTAYTCATAYTGATC | 25 | 52.0–54.9 |
| REV3 | As | CAGGATATTTATATTCATACTGGTC | 25 | 52.9 |
| Probe1 | S | Same as original (LN34 probe) [26] | 17 | 48.7–51.6 |
| Probe2 | S | Same as original (LN34lago probe) [26] | 17 | 45.8 |
| TNMM * | Phylogroup I (n = 251) | Phylogroup II (n = 12) | Phylogroup III (n = 6) | |||
|---|---|---|---|---|---|---|
| Primers | Probes | Primers | Probes | Primers | Probes | |
| 0 | 58% | 99% | 0% | 50% | 83% | 83% |
| 1 | 37% | 1% | 50% | 50% | 0% | 17% |
| 2 | 5% | 0% | 25% | 0% | 17% | 0% |
| 3 | 0% | 0% | 25% | 0% | 0% | 0% |
| 4 | 0% | 0% | 0% | 0% | 0% | 0% |
| 5 | 0% | 0% | 0% | 0% | 0% | 0% |
| 6 | 0% | 0% | 0% | 0% | 0% | 0% |
| 7 | 0% | 0% | 0% | 0% | 0% | 0% |
| AC** | 100% | 100% | 75% | 100% | 100% | 100% |
| Ph. * | RNA Transcript Dilution (GC/µL) | Within-Run Repeatability | Between-Days Repeatability | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | ||||||||||||
| Mean | SD | CV | % | Mean | SD | CV | % | Mean | SD | CV | % | ||
| I | EBLV-1 a—low (1 × 102) | 32.8 | 0.70 | 0.02 | 97.87 | 32.3 | 0.59 | 0.02 | 98.17 | 32.6 | 0.63 | 0.02 | 98.05 |
| EBLV-1 a—medium (1 × 104) | 24.6 | 0.25 | 0.01 | 99.00 | 24.8 | 0.26 | 0.01 | 98.97 | 24.7 | 0.25 | 0.01 | 99.00 | |
| II | LBV a—low (1 × 102) | 32.5 | 0.86 | 0.03 | 97.37 | 32.1 | 0.75 | 0.02 | 97.67 | 32.3 | 0.75 | 0.02 | 97.66 |
| LBV a—medium (1 × 104) | 25.8 | 0.84 | 0.03 | 96.72 | 26.7 | 0.03 | 0.00 | 99.89 | 26.3 | 0.75 | 0.03 | 97.15 | |
| III | IKOV—low (1 × 102) | 32.7 | 0.70 | 0.02 | 97.85 | 33.6 | 0.53 | 0.02 | 98.44 | 33.2 | 0.73 | 0.02 | 97.81 |
| IKOV—medium (1 × 104) | 24.8 | 0.51 | 0.02 | 97.96 | 25.0 | 0.02 | 0.00 | 99.91 | 24.9 | 0.34 | 0.01 | 98.63 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drzewnioková, P.; Marciano, S.; Leopardi, S.; Panzarin, V.; De Benedictis, P. Comparison of Pan-Lyssavirus RT-PCRs and Development of an Improved Protocol for Surveillance of Non-RABV Lyssaviruses. Viruses 2023, 15, 680. https://doi.org/10.3390/v15030680
Drzewnioková P, Marciano S, Leopardi S, Panzarin V, De Benedictis P. Comparison of Pan-Lyssavirus RT-PCRs and Development of an Improved Protocol for Surveillance of Non-RABV Lyssaviruses. Viruses. 2023; 15(3):680. https://doi.org/10.3390/v15030680
Chicago/Turabian StyleDrzewnioková, Petra, Sabrina Marciano, Stefania Leopardi, Valentina Panzarin, and Paola De Benedictis. 2023. "Comparison of Pan-Lyssavirus RT-PCRs and Development of an Improved Protocol for Surveillance of Non-RABV Lyssaviruses" Viruses 15, no. 3: 680. https://doi.org/10.3390/v15030680
APA StyleDrzewnioková, P., Marciano, S., Leopardi, S., Panzarin, V., & De Benedictis, P. (2023). Comparison of Pan-Lyssavirus RT-PCRs and Development of an Improved Protocol for Surveillance of Non-RABV Lyssaviruses. Viruses, 15(3), 680. https://doi.org/10.3390/v15030680





