Structure-Based Regulatory Role for the 5′UTR of RCNMV RNA2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viral Constructs
2.2. Preparation of Infectious Viral RNAs
2.3. SHAPE RNA Secondary Structure Analysis
2.4. Protoplast Transfections and Viral RNA and Protein Analysis
2.5. In Vitro Translation Assay
2.6. In Vitro Xrn1 Digestion Assay
3. Results
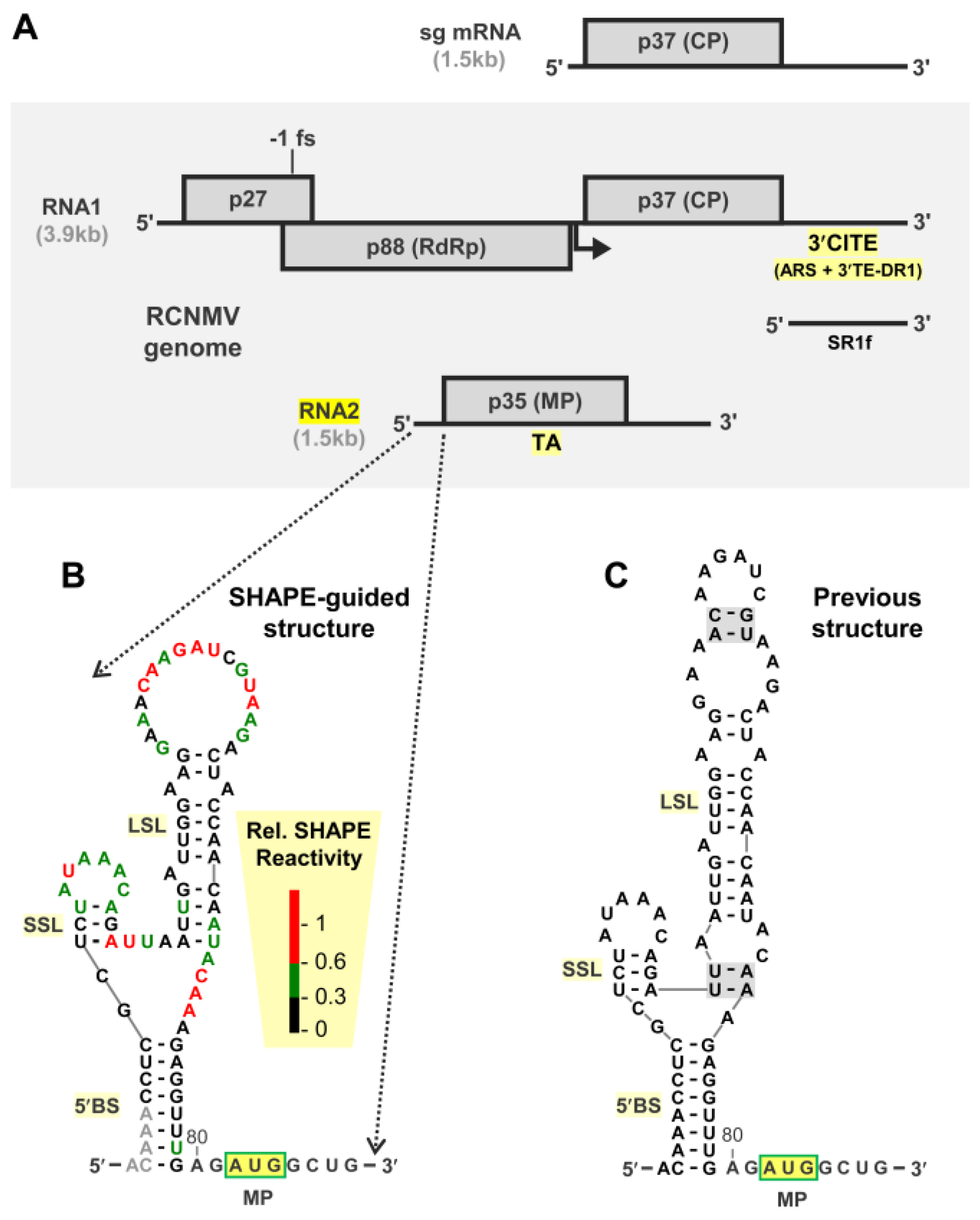
3.1. Structural Analysis of the 5′UTR of RNA2
3.2. RNA2 Translation Initiation Occurs through 5′-end Entry and Scanning
3.3. The 5′BS Is Required for RNA2 Replication and Inhibits Translation of MP
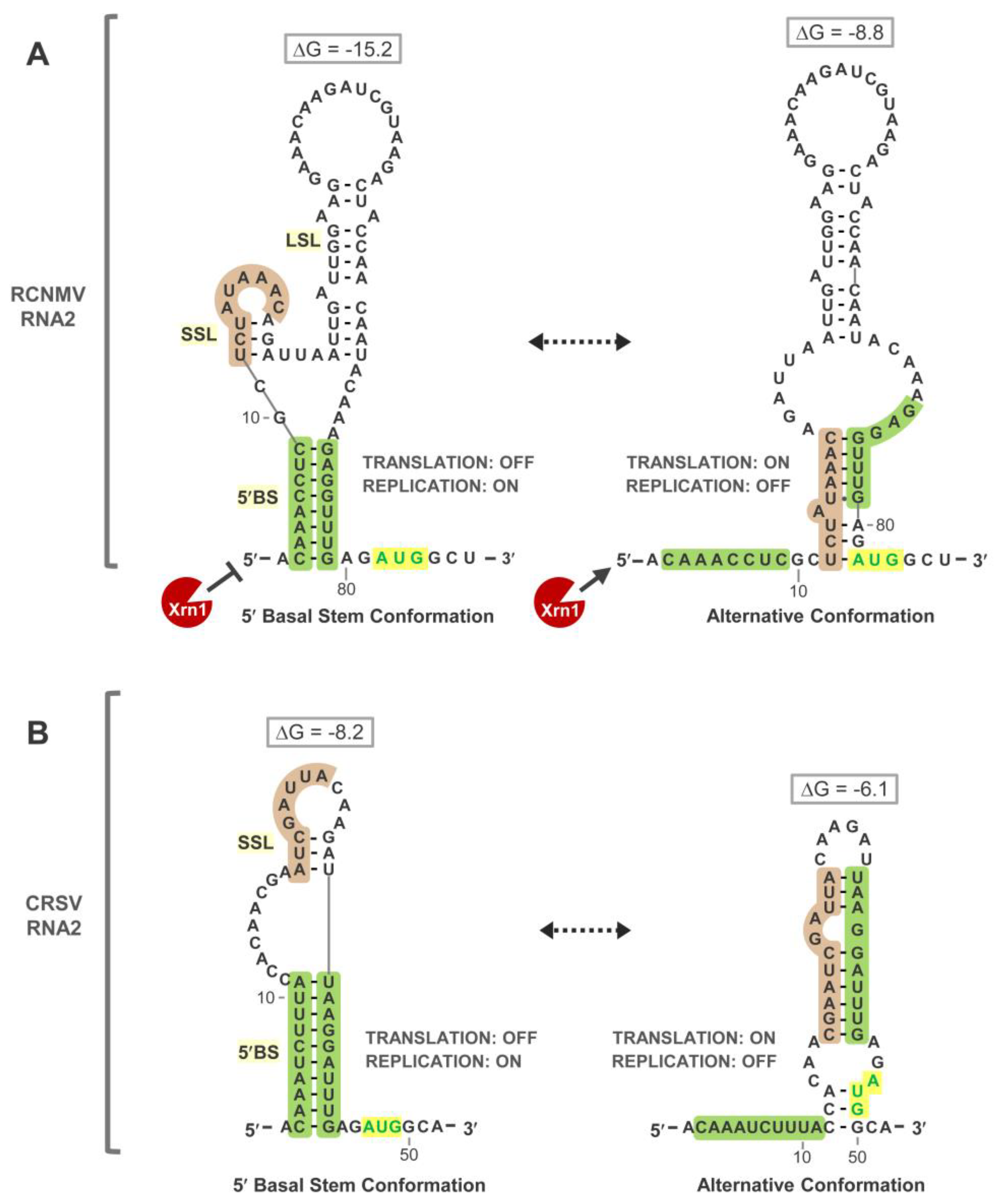
3.4. Exploring Alternative Conformations of the 5′UTR of RNA2
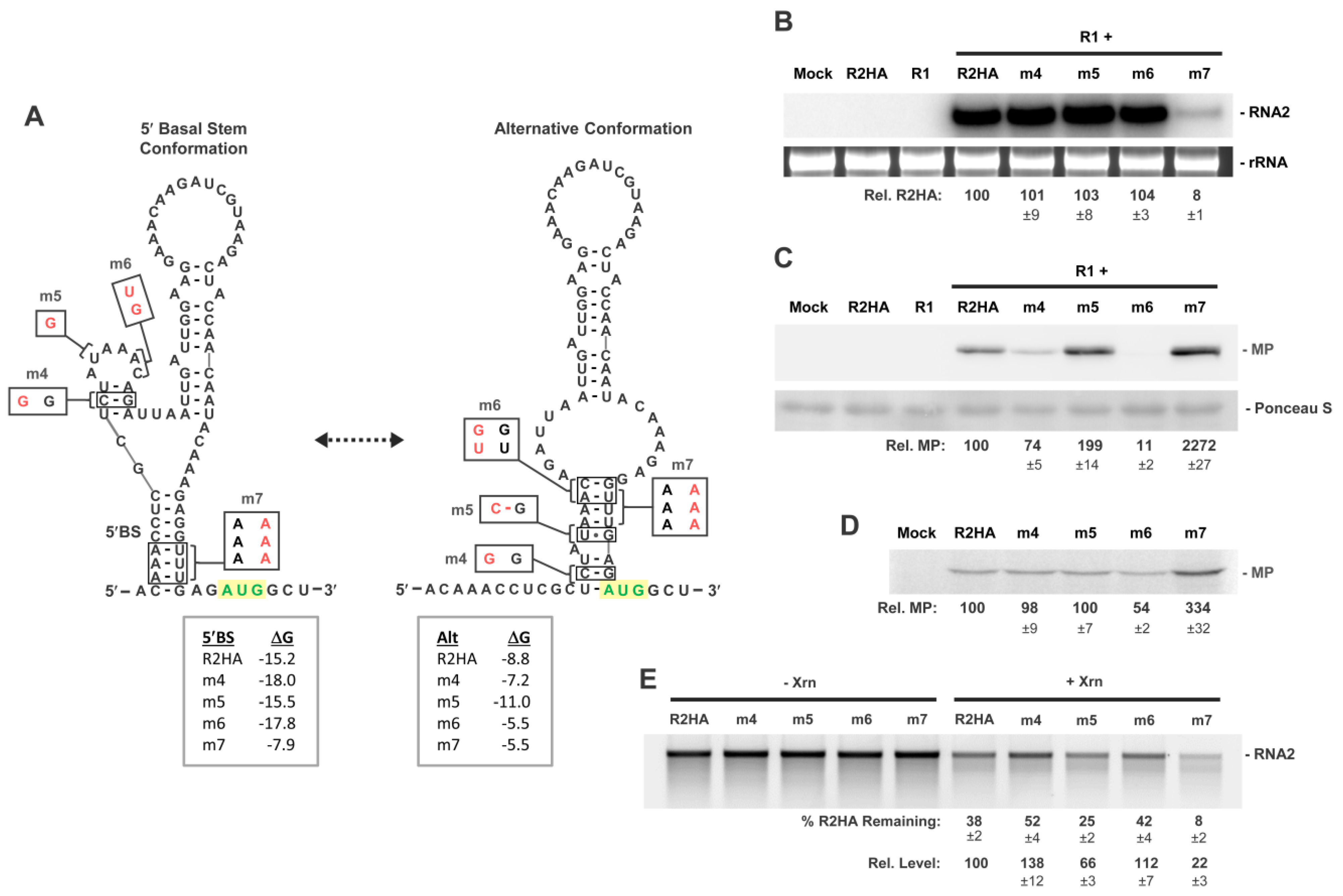
3.5. Assessing Functionality of RNA2 5′UTR Conformations
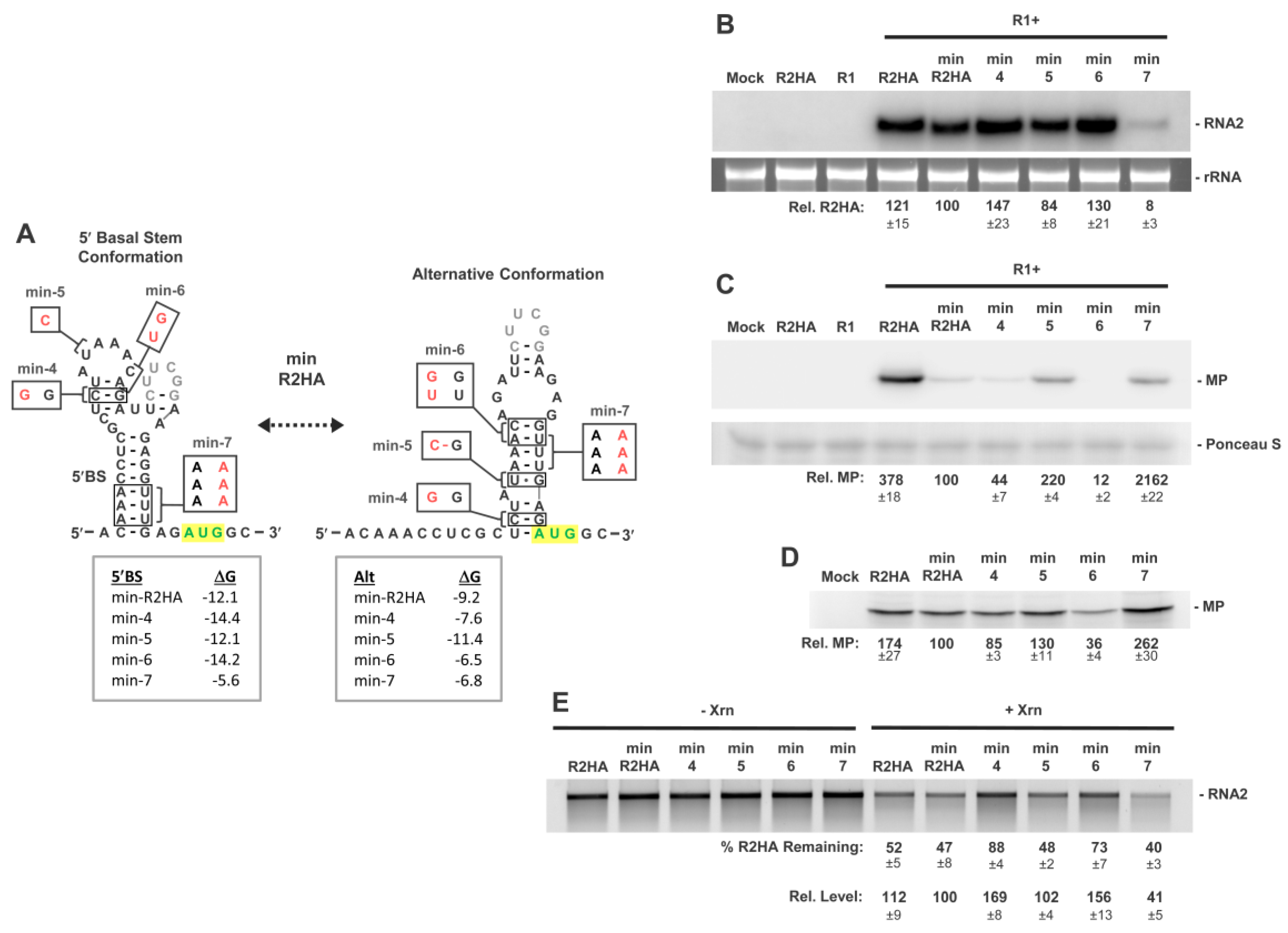
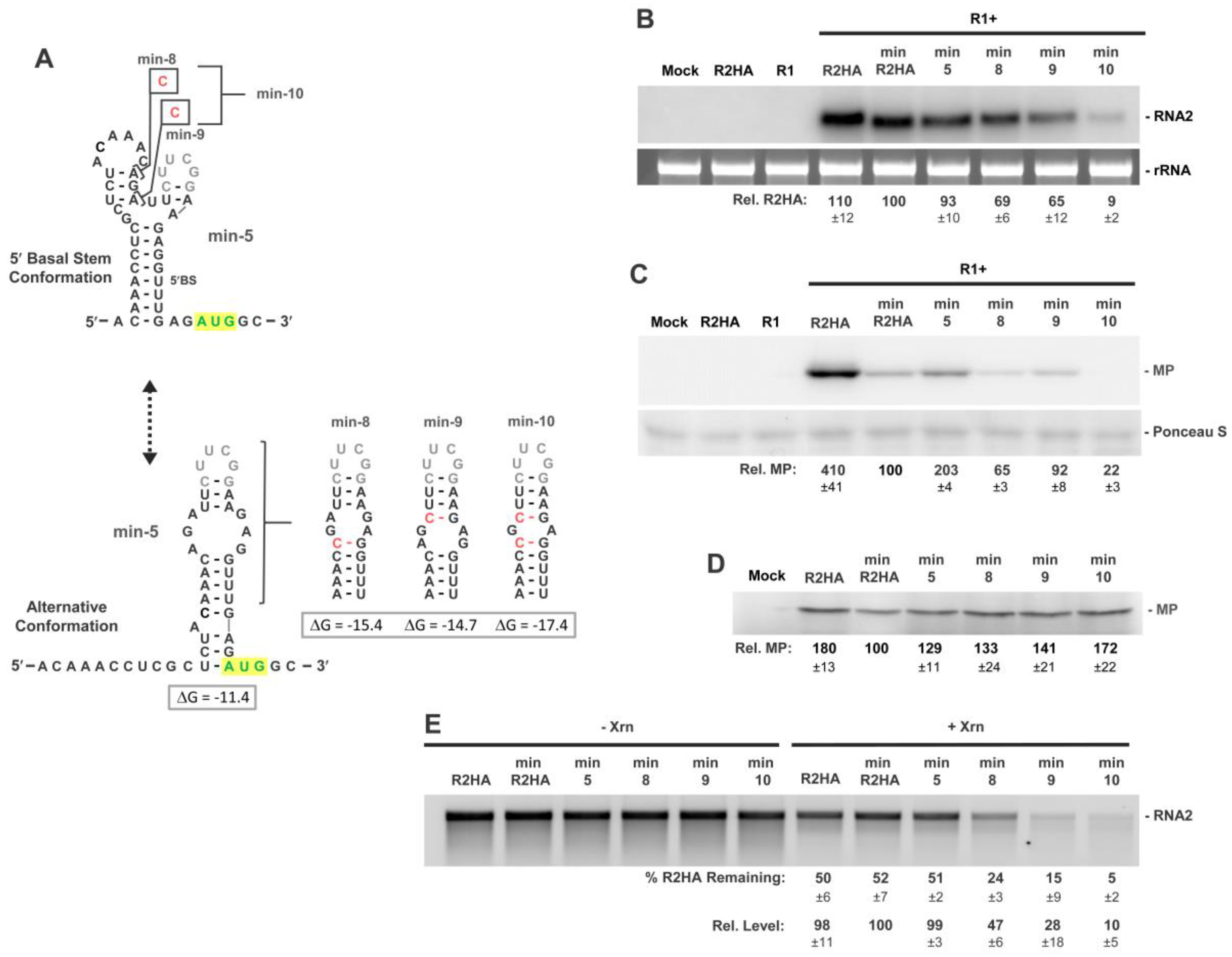
3.6. Activity of a Minimal 5′UTR in RNA2
3.7. Conformational Flexibility in the 5′UTR Is Required for Optimal RNA2 Activities
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gamarnik, A.V.; Andino, R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998, 12, 2293–2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimino, P.A.; Nicholson, B.L.; Wu, B.; Xu, W.; White, K.A. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog. 2011, 7, e1002423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Shi, K.; Meskauskas, A.; Simon, A.E. The 3′ end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA 2009, 15, 1849–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fajardo, T.; Sanford, T.J.; Mears, H.V.; Jasper, A.; Storrie, S.; Mansur, D.S.; Sweeney, T.R. The flavivirus polymerase NS5 regulates translation of viral genomic RNA. Nucleic Acids Res. 2020, 48, 5081–5093. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Z.; Lommel, S.A. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology 1989, 171, 543–554. [Google Scholar] [CrossRef]
- Okuno, T.; Hiruki, C. Molecular biology and epidemiology of dianthoviruses. Adv. Virus Res. 2013, 87, 37–74. [Google Scholar]
- Kusumanegara, K.; Mine, A.; Hyodo, K.; Kaido, M.; Mise, K.; Okuno, T. Identification of domains in p27 auxiliary replicase protein essential for its association with the endoplasmic reticulum membranes in red clover necrotic mosaic virus. Virology 2012, 433, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Takeda, A.; Taniguchi, T.; Taniguchi, H.; Kaido, M.; Mise, K.; Okuno, T. Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus. J. Virol. 2010, 84, 6070–6081. [Google Scholar] [CrossRef] [Green Version]
- Tajima, Y.; Iwakawa, H.O.; Kaido, M.; Mise, K.; Okuno, T. A long-distance RNA-RNA interaction plays an important role in programmed-1 ribosomal frameshifting in the translation of p88 replicase protein of Red clover necrotic mosaic virus. Virology 2011, 417, 169–178. [Google Scholar] [CrossRef] [Green Version]
- Sit, T.L.; Vaewhongs, A.A.; Lommel, S.A. RNA-mediated trans-activation of transcription from a viral RNA. Science 1998, 281, 829–832. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Mizumoto, H.; Nagano, H.; Imoto, Y.; Takigawa, K.; Sarawaneeyaruk, S.; Kaido, M.; Mise, K.; Okuno, T. A viral noncoding RNA generated by cis-element-mediated protection against 5′-3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 2008, 82, 10162–10174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanodia, P.; Miller, W.A. Effects of the noncoding subgenomic RNA of red clover necrotic mosaic virus in virus infection. J. Virol. 2022, 96, e0181521. [Google Scholar] [CrossRef] [PubMed]
- Steckelberg, A.L.; Vicens, Q.; Kieft, J.S. Exoribonuclease-resistant RNAs exist within both coding and noncoding subgenomic RNAs. mBio 2018, 9, e0246118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaido, M.; Funatsu, N.; Tsuno, Y.; Mise, K.; Okuno, T. Viral cell-to-cell movement requires formation of cortical punctate structures containing red clover necrotic mosaic virus movement protein. Virology 2011, 413, 205–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaido, M.; Abe, K.; Mine, A.; Hyodo, K.; Taniguchi, T.; Taniguchi, H.; Mise, K.; Okuno, T. GAPDH–a recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement. PLoS Pathog. 2014, 10, e1004505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatsuta, M.; Mizumoto, H.; Kaido, M.; Mise, K.; Okuno, T. The red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J. Virol. 2005, 79, 978–986. [Google Scholar] [CrossRef] [Green Version]
- An, M.; Iwakawa, H.O.; Mine, A.; Kaido, M.; Mise, K.; Okuno, T. A Y-shaped RNA structure in the 3′ untranslated region together with the trans-activator and core promoter of Red clover necrotic mosaic virus RNA2 is required for its negative-strand RNA synthesis. Virology 2010, 405, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Iwakawa, H.O.; Mine, A.; Hyodo, K.; An, M.; Kaido, M.; Mise, K.; Okuno, T. Template recognition mechanisms by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J. Virol. 2011, 85, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Newburn, L.R.; White, K.A. A trans-activator-like structure in RCNMV RNA1 evokes the origin of the trans-activator in RNA2. PLoS Pathog. 2020, 16, e1008271. [Google Scholar] [CrossRef]
- Mizumoto, H.; Tatsuta, M.; Kaido, M.; Mise, K.; Okuno, T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J. Virol. 2003, 77, 12113–12121. [Google Scholar] [CrossRef] [Green Version]
- Mizumoto, H.; Iwakawa, H.O.; Kaido, M.; Mise, K.; Okuno, T. Cap-independent translation mechanism of red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J. Virol. 2006, 80, 3781–3791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwakawa, H.O.; Kaido, M.; Mise, K.; Okuno, T. cis-Acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′-untranslated region of Red clover necrotic mosaic virus RNA1. Virology 2007, 369, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Sarawaneeyaruk, S.; Iwakawa, H.O.; Mizumoto, H.; Murakami, H.; Kaido, M.; Mise, K.; Okuno, T. Host-dependent roles of the viral 5′ untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3′ UTR of Red clover necrotic mosaic virus RNA1. Virology 2009, 391, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, J.S.H.; Newburn, L.R.; Kent, G.; White, K.A. Trans-activator binding site context in RCNMV modulates subgenomic mRNA transcription. Viruses 2021, 13, 2252. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tajima, Y.; Taniguchi, T.; Kaido, M.; Mise, K.; Tomari, Y.; Taniguchi, H.; Okuno, T. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3′ untranslated region. J. Virol. 2012, 86, 7836–7849. [Google Scholar] [CrossRef] [Green Version]
- Tajima, Y.; Iwakawa, H.O.; Hyodo, K.; Kaido, M.; Mise, K.; Okuno, T. Requirement for eukaryotic translation initiation factors in cap-independent translation differs between bipartite genomic RNAs of red clover necrotic mosaic virus. Virology 2017, 509, 152–158. [Google Scholar] [CrossRef]
- Hyodo, K.; Nagai, H.; Okuno, T. Dual function of a cis-acting RNA element that acts as a replication enhancer and a translation repressor in a plant positive-stranded RNA virus. Virology 2017, 512, 74–82. [Google Scholar] [CrossRef]
- White, K.A.; Morris, T.J. Nonhomologous RNA recombination in tombusviruses: Generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 1994, 68, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Gunawardene, C.D.; Newburn, L.R.; White, K.A. A 212-nt long RNA structure in the tobacco necrosis virus-D RNA genome is resistant to Xrn degradation. Nucleic Acids Res. 2019, 47, 9329–9342. [Google Scholar] [CrossRef]
- Low, J.T.; Weeks, K.M. SHAPE-directed RNA secondary structure prediction. Methods 2010, 52, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Vasa, S.M.; Guex, N.; Wilkinson, K.A.; Weeks, K.M.; Giddings, M.C. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information. RNA 2008, 14, 1979–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, P.Z.; Kasprzak, W.P.; Shapiro, B.A.; Simon, A.E. RNA2Drawer: Geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences. RNA Biol. 2019, 16, 1667–1671. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L.; White, K.A. Context-influenced cap-independent translation of Tombusvirus mRNAs in vitro. Virology 2008, 380, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Gunawardene, C.D.; Im, J.S.H.; White, K.A. RNA structure protects the 5′ end of an uncapped tombusvirus RNA genome from Xrn digestion. J. Virol. 2021, 95, e0103421. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Jinek, M.; Coyle, S.M.; Doudna, J.A. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell. 2011, 41, 600–608. [Google Scholar] [CrossRef] [Green Version]
- Langeberg, C.J.; Welch, W.R.W.; McGuire, J.V.; Ashby, A.; Jackson, A.D.; Chapman, E.G. Biochemical characterization of yeast Xrn1. Biochemistry 2020, 59, 1493–1507. [Google Scholar] [CrossRef]
- Kendall, T.L.; Lommel, S.A. Nucleotide sequence of carnation ringspot dianthovirus RNA-2. J. Gen. Virol. 1992, 73, 2479–2482. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Generozov, E.V.; Kendall, T.L.; Lommel, S.A.; Zavriev, S.K. Nucleotide sequence of carnation ringspot dianthovirus RNA-1. J. Gen. Virol. 1994, 75, 243–247. [Google Scholar] [CrossRef]
- Kaido, M.; Nagano, H.; Omote, K.; Takano, Y.; Mise, K.; Okuno, T. 5′-Terminal stem-loop of carnation ringspot virus RNA1 is required for the efficient amplification of viral RNAs. Virus Res. 2019, 265, 138–142. [Google Scholar] [CrossRef]
- Turner, R.L.; Buck, K.W. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology 1999, 253, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Z.; Xiong, Z. Three discontinuous loop nucleotides in the 3′ terminal stem-loop are required for Red clover necrotic mosaic virus RNA-2 replication. Virology 2009, 393, 346–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, G.; Xin, S.; Cai, Z. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 2003, 77, 3312–3318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallie, D.R.; Browning, K.S. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 2001, 276, 36951–36960. [Google Scholar] [CrossRef] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Im, J.S.H.; Sheppard, J.R.; White, K.A. Structure-Based Regulatory Role for the 5′UTR of RCNMV RNA2. Viruses 2023, 15, 722. https://doi.org/10.3390/v15030722
Im JSH, Sheppard JR, White KA. Structure-Based Regulatory Role for the 5′UTR of RCNMV RNA2. Viruses. 2023; 15(3):722. https://doi.org/10.3390/v15030722
Chicago/Turabian StyleIm, Jennifer S. H., Jasmine R. Sheppard, and K. Andrew White. 2023. "Structure-Based Regulatory Role for the 5′UTR of RCNMV RNA2" Viruses 15, no. 3: 722. https://doi.org/10.3390/v15030722
APA StyleIm, J. S. H., Sheppard, J. R., & White, K. A. (2023). Structure-Based Regulatory Role for the 5′UTR of RCNMV RNA2. Viruses, 15(3), 722. https://doi.org/10.3390/v15030722






