HLA Variation and SARS-CoV-2 Specific Antibody Response
Abstract
:1. Introduction
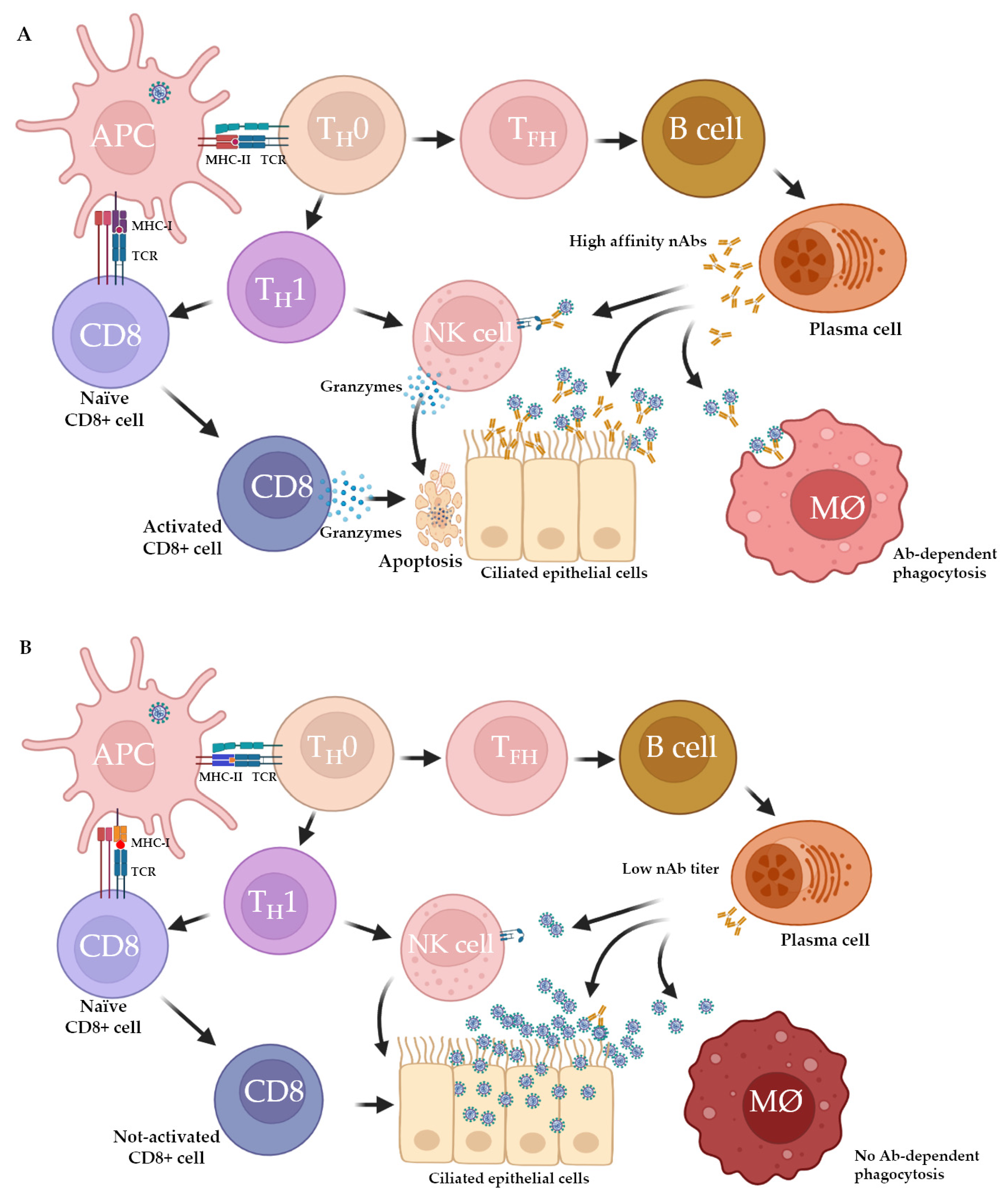
2. Host Immune Response to SARS-CoV-2: Innate and Adaptive Immunity
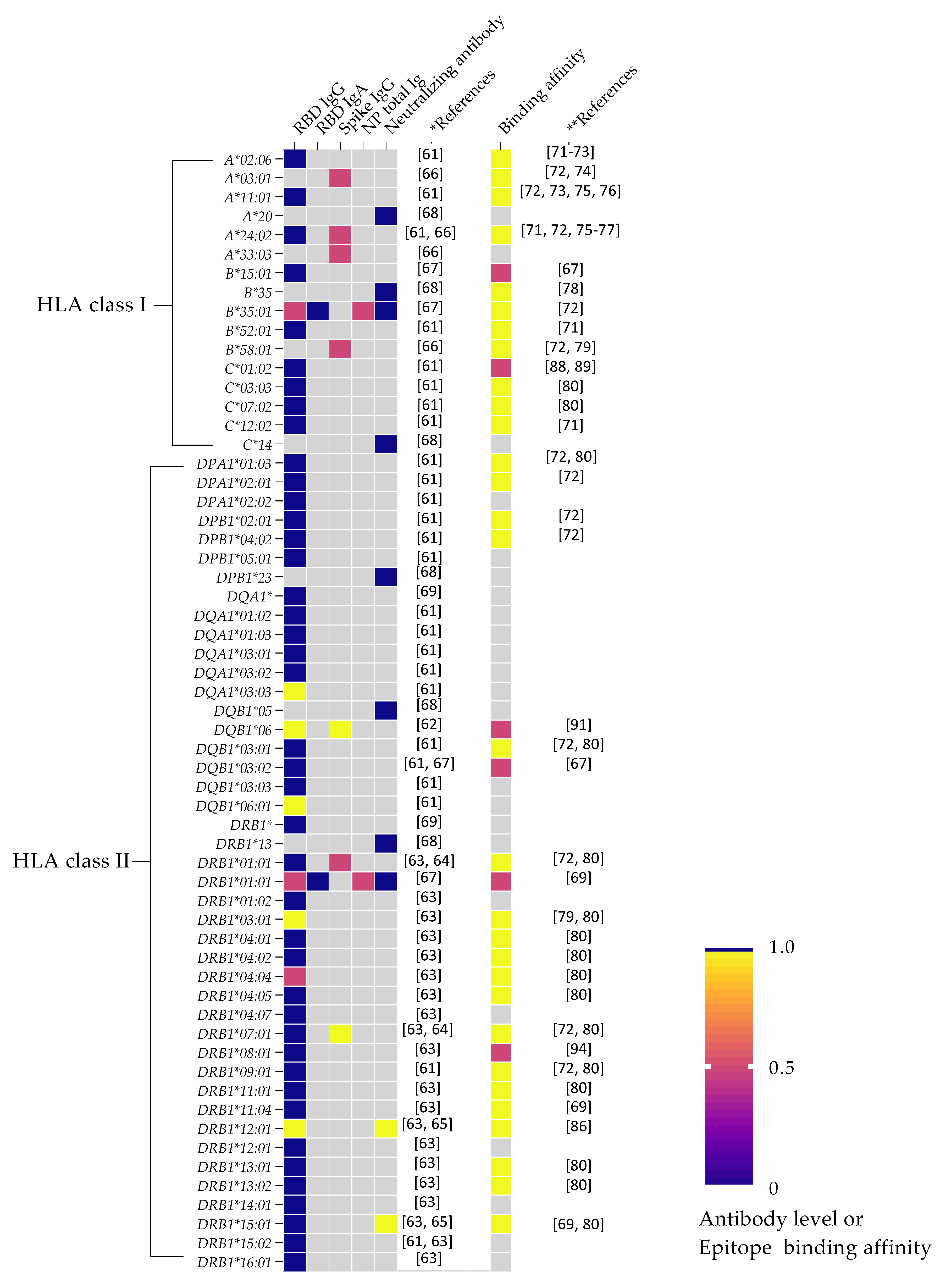
3. HLA Variation and Humoral Immunity in SARS-CoV-2
4. SARS-CoV-2 Peptide Epitope Binding Affinity and Antibody Response
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- O’Driscoll, M.; Dos Santos, G.R.; Wang, L.; Cummings, D.A.T.; Azman, A.S.; Paireau, J.; Fontanet, A.; Cauchemez, S.; Salje, H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 2021, 590, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; Tokuyama, M.; et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020, 588, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Kullar, R.; Marcelin, J.R.; Swartz, T.H.; Piggott, D.A.; Macias Gil, R.; Mathew, T.A.; Tan, T. Racial disparity of Coronavirus Disease 2019 in African American communities. J. Infect. Dis. 2020, 222, 890–893. [Google Scholar] [CrossRef]
- Navaratnam, A.V.; Gray, W.K.; Day, J.; Wendon, J.; Briggs, T.W.R. Patient factors and temporal trends associated with COVID-19 in-hospital mortality in England: An observational study using administrative data. Lancet Respir. Med. 2021, 9, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.D.; Moffitt, R.A.; Hajagos, J.G.; Amor, B.; Anand, A.; Bissell, M.M.; Bradwell, K.R.; Bremer, C.; Byrd, J.B.; Denham, A.; et al. Clinical Characterization and Prediction of Clinical Severity of SARS-CoV-2 Infection Among US Adults Using Data from the US National COVID Cohort Collaborative. JAMA Netw. Open 2021, 4, e2116901. [Google Scholar] [CrossRef] [PubMed]
- Wolday, D.; Gebrecherkos, T.; Arefaine, Z.G.; Kiros, Y.K.; Gebreegzabher, A.; Tasew, G.; Abdulkader, M.; Abraha, H.E.; Desta, A.A.; Hailu, A.; et al. Effect of co-infection with intestinal parasites on COVID-19 severity: A prospective observational cohort study. eClinicalMedicine 2021, 39, 101054. [Google Scholar] [CrossRef]
- Lauring, A.S.; Tenforde, M.W.; Chappell, J.D.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Clinical severity of, and effectiveness of mRNA vaccines against, COVID-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: Prospective observational study. BMJ 2022, 376, e069761. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Karbuz, A.; Gervais, A.; Tayoun, A.A.; Aiuti, A.; Belot, A.; Bolze, A.; Gaudet, A.; Bondarenko, A.; et al. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 2022, 603, 587–598. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Petersen, J.; Rossjohn, J.; Fugger, L. HLA variation and disease. Nat. Rev. Immunol. 2018, 18, 325–339. [Google Scholar] [CrossRef]
- Stephens, H.A.F.; Klaythong, R.; Sirikong, M.; Vaughn, D.W.; Green, S.; Kalayanarooj, S.; Endy, T.P.; Libraty, D.H.; Nisalak, A.; Innis, B.; et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens 2002, 60, 309–318. [Google Scholar] [CrossRef]
- Jiang, Y.-G.; Wang, Y.-M.; Liu, T.-H.; Liu, J. Association between HLA class II gene and susceptibility or resistance to chronic hepatitis B. World J. Gastroenterol. 2003, 9, 2221–2225. [Google Scholar] [CrossRef] [PubMed]
- Luo, M. Natural Immunity against HIV-1: Progression of Understanding after Association Studies. Viruses 2022, 14, 1243. [Google Scholar] [CrossRef] [PubMed]
- Harishankar, M.; Selvaraj, P.; Bethunaickan, R. Influence of Genetic Polymorphism Towards Pulmonary Tuberculosis Susceptibility. Front. Med. 2018, 5, 213–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kano, F.S.; Souza-Silva, F.A.; Torres, L.M.; Lima, B.A.S.; Sousa, T.N.; Alves, J.R.S.; Rocha, R.S.; Fontes, C.J.F.; Sanchez, B.A.M.; Adams, J.H.; et al. The Presence, Persistence and Functional Properties of Plasmodium vivax Duffy Binding Protein II Antibodies Are Influenced by HLA Class II Allelic Variants. PLoS Negl. Trop. Dis. 2016, 10, e0005177. [Google Scholar] [CrossRef] [Green Version]
- Ovsyannikova, I.G.; Pankratz, V.S.; Vierkant, R.A.; Jacobson, R.M.; Poland, G.A. Human Leukocyte Antigen Haplotypes in the Genetic Control of Immune Response to Measles-Mumps-Rubella Vaccine. J. Infect. Dis. 2006, 193, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ovsyannikova, I.G.; Vierkant, R.A.; Pankratz, V.S.; Jacobson, R.M.; Poland, G.A. Human Leukocyte Antigen Genotypes in the Genetic Control of Adaptive Immune Responses to Smallpox Vaccine. J. Infect. Dis. 2011, 203, 1546–1555. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, T.; Yu, Q.; Zhang, H.; Du, J.; Zhang, Y.; Xia, S.; Yang, H.; Li, Q. Association of human leukocyte antigen alleles and supertypes with immunogenicity of oral rotavirus vaccine given to infants in China. Medicine 2018, 97, e12706. [Google Scholar] [CrossRef]
- Png, E.; Thalamuthu, A.; Ong, R.T.; Snippe, H.; Boland, G.J.; Seielstad, M. A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Hum. Mol. Genet. 2011, 20, 3893–3898. [Google Scholar] [CrossRef]
- Pan, L.; Zhang, L.; Zhang, W.; Wu, X.; Li, Y.; Yan, B.; Zhu, X.; Liu, X.; Yang, C.; Xu, J.; et al. A genome-wide association study identifies polymorphisms in the HLA-DR region associated with non-response to hepatitis B vaccination in Chinese Han populations. Hum. Mol. Genet. 2013, 23, 2210–2219. [Google Scholar] [CrossRef] [Green Version]
- Nishida, N.; Sugiyama, M.; Sawai, H.; Nishina, S.; Sakai, A.; Ohashi, J.; Khor, S.; Kakisaka, K.; Tsuchiura, T.; Hino, K.; et al. Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology 2018, 68, 848–858. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.; Roh, E.Y.; Park, B.; Lee, Y.; Shin, S.; Yoon, J.H.; Song, E.Y. GWAS identifying HLA-DPB1 gene variants associated with responsiveness to hepatitis B virus vaccination in Koreans: Independent association of HLA-DPB1*04:02 possessing rs1042169 G-rs9277355 C-rs9277356 A. J. Viral Hepat. 2019, 26, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Wei, H.; Li, M.; Cheng, Y.; Wen, S.; Wang, D.; Shu, Y. Single Nucleotide Polymorphisms in the Human Leukocyte Antigen Region Are Associated with Hemagglutination Inhibition Antibody Response to Influenza Vaccine. Front. Genet. 2022, 13, 790914. [Google Scholar] [CrossRef] [PubMed]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.E.K.; Daly, M.J.; Ganna, A. The human genetic epidemiology of COVID-19. Nat. Rev. Genet. 2022, 23, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Zepeda-Cervantes, J.; Martínez-Flores, D.; Ramírez-Jarquín, J.O.; Tecalco-Cruz, C.; Alavez-Pérez, N.S.; Vaca, L.; Sarmiento-Silva, R.E. Implications of the Immune Polymorphisms of the Host and the Genetic Variability of SARS-CoV-2 in the Development of COVID-19. Viruses 2022, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Carsetti, R.; Zaffina, S.; Mortari, E.P.; Terreri, S.; Corrente, F.; Capponi, C.; Palomba, P.; Mirabella, M.; Cascioli, S.; Palange, P.; et al. Different Innate and Adaptive Immune Responses to SARS-CoV-2 Infection of Asymptomatic, Mild, and Severe Cases. Front. Immunol. 2020, 11, 610300. [Google Scholar] [CrossRef]
- Hall, V.; Foulkes, S.; Insalata, F.; Kirwan, P.; Saei, A.; Atti, A.; Wellington, E.; Khawam, J.; Munro, K.; Cole, M.; et al. Protection against SARS-CoV-2 after Covid-19 Vaccination and Previous Infection. N. Engl. J. Med. 2022, 386, 1207–1220. [Google Scholar] [CrossRef]
- Rose, R.; Neumann, F.; Grobe, O.; Lorentz, T.; Fickenscher, H.; Krumbholz, A. Humoral immune response after different SARS-CoV-2 vaccination regimens. BMC Med. 2022, 20, 31. [Google Scholar] [CrossRef]
- Barin, B.; Kasap, U.; Selçuk, F.; Volkan, E.; Uluçkan, O. Comparison of SARS-CoV-2 anti-spike receptor binding domain IgG antibody responses after CoronaVac, BNT162b2, ChAdOx1 COVID-19 vaccines, and a single booster dose: A prospective, longitudinal population-based study. Lancet Microbe 2022, 3, e274–e283. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; Macary, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2020, 22, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Nagler, A.; Kalaora, S.; Barbolin, C.; Gangaev, A.; Ketelaars, S.L.; Alon, M.; Pai, J.; Benedek, G.; Yahalom-Ronen, Y.; Erez, N.; et al. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep. 2021, 35, 109305. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Linster, M.; Tan, C.W.; Le Bert, N.; Ni Chia, W.; Kunasegaran, K.; Zhuang, Y.; Tham, C.Y.L.; Chia, A.; Smith, G.J.D.; et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021, 34, 108728. [Google Scholar] [CrossRef] [PubMed]
- Juno, J.A.; Tan, H.-X.; Lee, W.S.; Reynaldi, A.; Kelly, H.G.; Wragg, K.; Esterbauer, R.; Kent, H.E.; Batten, C.J.; Mordant, F.L.; et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020, 26, 1428–1434. [Google Scholar] [CrossRef]
- Boppana, S.; Qin, K.; Files, J.K.; Russell, R.M.; Stoltz, R.; Bibollet-Ruche, F.; Bansal, A.; Erdmann, N.; Hahn, B.H.; Goepfert, P.A. SARS-CoV-2-specific circulating T follicular helper cells correlate with neutralizing antibodies and increase during early convalescence. PLoS Pathog. 2021, 17, e1009761. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Suthar, M.S.; Zimmerman, M.; Kauffman, R.; Mantus, G.; Linderman, S.; Vanderheiden, A.; Nyhoff, L.; Davis, C.; Adekunle, S.; Affer, M.; et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Altmann, D.M.; Boyton, R.J. SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Sci. Immunol. 2020, 5, eabd6160. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.C.; Lamothe, P.A.; Woodruff, M.C.; Saini, A.S.; Faliti, C.E.; Sanz, I.; Lee, F.E. COVID-19 and plasma cells: Is there long-lived protection? Immunol. Rev. 2022, 309, 40–63. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Juno, J.A.; Khoury, D.; Reynaldi, A.; Wheatley, A.K.; Kent, S.J.; Davenport, M.P. Prospects for durable immune control of SARS-CoV-2 and prevention of reinfection. Nat. Rev. Immunol. 2021, 21, 395–404. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Reusch, J.; Wagenhäuser, I.; Gabel, A.; Eggestein, A.; Höhn, A.; Lâm, T.; Frey, A.; Schubert-Unkmeir, A.; Dölken, L.; Frantz, S.; et al. Influencing factors of anti-SARS-CoV-2-spike-IgG antibody titers in healthcare workers: A cross-section study. J. Med. Virol. 2022, 95, e28300. [Google Scholar] [CrossRef]
- Karachaliou, M.; Moncunill, G.; Espinosa, A.; Castaño-Vinyals, G.; Rubio, R.; Vidal, M.; Jiménez, A.; Prados, E.; Carreras, A.; Cortés, B.; et al. SARS-CoV-2 infection, vaccination, and antibody response trajectories in adults: A cohort study in Catalonia. BMC Med. 2022, 20, 347. [Google Scholar] [CrossRef]
- Lindemann, M.; Klisanin, V.; Thümmler, L.; Fisenkci, N.; Tsachakis-Mück, N.; Ditschkowski, M.; Schwarzkopf, S.; Klump, H.; Reinhardt, H.C.; Horn, P.A.; et al. Humoral and Cellular Vaccination Responses against SARS-CoV-2 in Hematopoietic Stem Cell Transplant Recipients. Vaccines 2021, 9, 1075. [Google Scholar] [CrossRef]
- Dayam, R.M.; Law, J.C.; Goetgebuer, R.L.; Chao, G.Y.; Abe, K.T.; Sutton, M.; Finkelstein, N.; Stempak, J.M.; Pereira, D.; Croitoru, D.; et al. Accelerated waning of immunity to SARS-CoV-2 mRNA vaccines in patients with immune-mediated inflammatory diseases. J. Clin. Investig. 2022, 7, e159721. [Google Scholar] [CrossRef]
- Moncunill, G.; Aguilar, R.; Ribes, M.; Ortega, N.; Rubio, R.; Salmerón, G.; Molina, M.J.; Vidal, M.; Barrios, D.; Mitchell, R.A.; et al. Determinants of early antibody responses to COVID-19 mRNA vaccines in a cohort of exposed and naïve healthcare workers. eBioMedicine 2022, 75, 103805. [Google Scholar] [CrossRef] [PubMed]
- Röltgen, K.; Powell, A.E.; Wirz, O.F.; Stevens, B.A.; Hogan, C.A.; Najeeb, J.; Hunter, M.; Wang, H.; Sahoo, M.K.; Huang, C.; et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020, 5, eabe0240. [Google Scholar] [CrossRef] [PubMed]
- Błaszczuk, A.; Michalski, A.; Sikora, D.; Malm, M.; Drop, B.; Polz-Dacewicz, M. Antibody Response after SARS-CoV-2 Infection with the Delta and Omicron Variant. Vaccines 2022, 10, 1728. [Google Scholar] [CrossRef] [PubMed]
- Medits, I.; Springer, D.N.; Graninger, M.; Camp, J.V.; Höltl, E.; Aberle, S.W.; Traugott, M.T.; Hoepler, W.; Deutsch, J.; Lammel, O.; et al. Different Neutralization Profiles After Primary SARS-CoV-2 Omicron BA.1 and BA.2 Infections. Front. Immunol. 2022, 13, 946318. [Google Scholar] [CrossRef] [PubMed]
- Uprichard, S.L.; O’Brien, A.; Evdokimova, M.; Rowe, C.L.; Joyce, C.; Hackbart, M.; Cruz-Pulido, Y.E.; Cohen, C.A.; Rock, M.L.; Dye, J.M.; et al. Antibody Response to SARS-CoV-2 Infection and Vaccination in COVID-19-naïve and Experienced Individuals. Viruses 2022, 14, 370. [Google Scholar] [CrossRef] [PubMed]
- Zheutlin, A.; Ott, M.; Sun, R.; Zemlianskaia, N.; Meyer, C.S.; Rubel, M.; Hayden, J.; Neri, B.; Kamath, T.; Khan, N.; et al. Durability of Protection Post–Primary COVID-19 Vaccination in the United States. Vaccines 2022, 10, 1458. [Google Scholar] [CrossRef]
- Bates, T.A.; McBride, S.K.; Leier, H.C.; Guzman, G.; Lyski, Z.L.; Schoen, D.; Winders, B.; Lee, J.-Y.; Lee, D.X.; Messer, W.B.; et al. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci. Immunol. 2022, 7, eabn8014. [Google Scholar] [CrossRef]
- Reynolds, C.J.; Pade, C.; Gibbons, J.M.; Butler, D.K.; Otter, A.D.; Menacho, K.; Fontana, M.; Smit, A.; Sackville-West, J.E.; Cutino-Moguel, T.; et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science 2021, 372, 1418–1423. [Google Scholar] [CrossRef]
- Olmstead, A.D.; Nikiforuk, A.M.; Schwartz, S.; Márquez, A.C.; Valadbeigy, T.; Flores, E.; Saran, M.; Goldfarb, D.M.; Hayden, A.; Masud, S.; et al. Characterizing Longitudinal Antibody Responses in Recovered Individuals Following COVID-19 Infection and Single-Dose Vaccination: A Prospective Cohort Study. Viruses 2022, 14, 2416. [Google Scholar] [CrossRef]
- Khor, S.-S.; Omae, Y.; Takeuchi, J.S.; Fukunaga, A.; Yamamoto, S.; Tanaka, A.; Matsuda, K.; Kimura, M.; Maeda, K.; Ueda, G.; et al. An Association Study of HLA with the Kinetics of SARS-CoV-2 Spike Specific IgG Antibody Responses to BNT162b2 mRNA Vaccine. Vaccines 2022, 10, 563. [Google Scholar] [CrossRef]
- Mentzer, A.J.; O’Connor, D.; Bibi, S.; Chelysheva, I.; Clutterbuck, E.A.; Demissie, T.; Dinesh, T.; Edwards, N.J.; Felle, S.; Feng, S.; et al. Human leukocyte antigen alleles associate with COVID-19 vaccine immunogenicity and risk of breakthrough infection. Nat. Med. 2022, 29, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Astbury, S.; Reynolds, C.J.; Butler, D.K.; Muñoz-Sandoval, D.C.; Lin, K.; Pieper, F.P.; Otter, A.; Kouraki, A.; Cusin, L.; Nightingale, J.; et al. HLA-DR polymorphism in SARS-CoV-2 infection and susceptibility to symptomatic COVID-19. Immunology 2022, 166, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bautista, J.F.; Sampedro, A.; Gómez-Vicente, E.; Rodríguez-Granger, J.; Reguera, J.A.; Cobo, F.; Ruiz-Cabello, F.; López-Nevot, M. HLA Class II Polymorphism and Humoral Immunity Induced by the SARS-CoV-2 mRNA-1273 Vaccine. Vaccines 2022, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Oka, S.; Furukawa, H.; Tohma, S. Associations of HLA Polymorphisms with Anti-SARS-CoV-2 Spike and Neutralizing Antibody Titers in Japanese Rheumatoid Arthritis Patients Vaccinated with BNT162b2. Vaccines 2023, 11, 404. [Google Scholar] [CrossRef]
- Crocchiolo, R.; Gallina, A.M.; Pani, A.; Campisi, D.; Cento, V.; Sacchi, N.; Miotti, V.; Gagliardi, O.M.; D’Amico, F.; Vismara, C.; et al. Polymorphism of the HLA system and weak antibody response to BNT162b2 mRNA vaccine. HLA 2022, 99, 183–191. [Google Scholar] [CrossRef]
- Fischer, J.C.; Schmidt, A.G.; Bölke, E.; Uhrberg, M.; Keitel, V.; Feldt, T.; Jensen, B.; Häussinger, D.; Adams, O.; Schneider, E.M.; et al. Association of HLA genotypes, AB0 blood type and chemokine receptor 5 mutant CD195 with the clinical course of COVID-19. Eur. J. Med. Res. 2021, 26, 107. [Google Scholar] [CrossRef]
- Weidner, L.; Kalser, J.; Kreil, T.R.; Jungbauer, C.; Mayr, W.R. Neutralizing Antibodies against SARS-CoV-2 and HLA Class I and II Polymorphism. Transfus. Med. Hemother. 2021, 48, 173–174. [Google Scholar] [CrossRef]
- Ragone, C.; Meola, S.; Fiorillo, P.C.; Penta, R.; Auriemma, L.; Tornesello, M.L.; Miscio, L.; Cavalcanti, E.; Botti, G.; Buonaguro, F.M.; et al. HLA Does Not Impact on Short-Medium-Term Antibody Response to Preventive Anti-SARS-Cov-2 Vaccine. Front. Immunol. 2021, 12, 734689. [Google Scholar] [CrossRef]
- Gerhards, C.; Kittel, M.; Ast, V.; Bugert, P.; Froelich, M.F.; Hetjens, M.; Haselmann, V.; Neumaier, M.; Thiaucourt, M. Humoral SARS-CoV-2 Immune Response in COVID-19 Recovered Vaccinated and Unvaccinated Individuals Related to Post-COVID-Syndrome. Viruses 2023, 15, 454. [Google Scholar] [CrossRef]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef]
- Romero-López, J.P.; Carnalla-Cortés, M.; Pacheco-Olvera, D.L.; Ocampo-Godínez, J.M.; Oliva-Ramírez, J.; Moreno-Manjón, J.; Bernal-Alferes, B.; López-Olmedo, N.; García-Latorre, E.; Domínguez-López, M.L.; et al. A bioinformatic prediction of antigen presentation from SARS-CoV-2 spike protein revealed a theoretical correlation of HLA-DRB1*01 with COVID-19 fatality in Mexican population: An ecological approach. J. Med. Virol. 2020, 93, 2029–2038. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef]
- Shkurnikov, M.; Nersisyan, S.; Jankevic, T.; Galatenko, A.; Gordeev, I.; Vechorko, V.; Tonevitsky, A. Association of HLA Class I Genotypes with Severity of Coronavirus Disease-19. Front. Immunol. 2021, 12, 641900. [Google Scholar] [CrossRef]
- Tomita, Y.; Ikeda, T.; Sato, R.; Sakagami, T. Association between HLA gene polymorphisms and mortality of COVID-19: An in silico analysis. Immun. Inflamm. Dis. 2020, 8, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Pretti, M.A.M.; Galvani, R.G.; Vieira, G.F.; Bonomo, A.; Bonamino, M.H.; Boroni, M. Class I HLA Allele Predicted Restricted Antigenic Coverages for Spike and Nucleocapsid Proteins Are Associated with Deaths Related to COVID-19. Front. Immunol. 2020, 11, 565730. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T. Human Leukocyte Antigen (HLA) Class I Susceptible Alleles Against COVID-19 Increase Both Infection and Severity Rate. Cureus 2020, 12, e12239. [Google Scholar] [CrossRef]
- Naemi, F.M.A.; Al-Adwani, S.; Al-Khatabi, H.; Al-Nazawi, A. Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J. Med. Virol. 2021, 93, 4430–4437. [Google Scholar] [CrossRef] [PubMed]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front. Immunol. 2020, 11, 605688. [Google Scholar] [CrossRef]
- Yarmarkovich, M.; Warrington, J.M.; Farrel, A.; Maris, J.M. Identification of SARS-CoV-2 Vaccine Epitopes Predicted to Induce Long-Term Population-Scale Immunity. Cell Rep. Med. 2020, 1, 100036. [Google Scholar] [CrossRef]
- Lorente, L.; Martín, M.; Franco, A.; Barrios, Y.; Cáceres, J.; Solé-Violán, J.; Perez, A.; Ramos, J.M.Y.; Ramos-Gómez, L.; Ojeda, N.; et al. HLA genetic polymorphisms and prognosis of patients with COVID-19. Med. Intensiv. 2021, 45, 96–103. [Google Scholar] [CrossRef]
- Wang, F.; Huang, S.; Gao, R.; Zhou, Y.; Lai, C.; Li, Z.; Xian, W.; Qian, X.; Li, Z.; Huang, Y.; et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Khor, S.-S.; Omae, Y.; Nishida, N.; Sugiyama, M.; Kinoshita, N.; Suzuki, T.; Suzuki, M.; Suzuki, S.; Izumi, S.; Hojo, M.; et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, Age and Sex Are Associated with Severity of Japanese COVID-19 with Respiratory Failure. Front. Immunol. 2021, 12, 658570. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Birol, I. Retrospective in silico HLA predictions from COVID-19 patients reveal alleles associated with disease prognosis. MedRxiv 2020. [Google Scholar] [CrossRef]
- Sakuraba, A.; Haider, H.; Sato, T. Population Difference in Allele Frequency of HLA-C*05 and Its Correlation with COVID-19 Mortality. Viruses 2020, 12, 1333. [Google Scholar] [CrossRef]
- Pisanti, S.; Deelen, J.; Gallina, A.M.; Caputo, M.; Citro, M.; Abate, M.; Sacchi, N.; Vecchione, C.; Martinelli, R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of COVID-19. J. Transl. Med. 2020, 18, 352. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Birol, I. HLA predictions from the bronchoalveolar lavage fluid and blood samples of eight COVID-19 patients at the pandemic onset. Bioinformatics 2020, 36, 5271–5273. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef] [Green Version]
- Correale, P.; Mutti, L.; Pentimalli, F.; Baglio, G.; Saladino, R.; Sileri, P.; Giordano, A. HLA-B*44 and C*01 Prevalence Correlates with Covid19 Spreading across Italy. Int. J. Mol. Sci. 2020, 21, 5205. [Google Scholar] [CrossRef]
- Novelli, A.; Andreani, M.; Biancolella, M.; Liberatoscioli, L.; Passarelli, C.; Colona, V.L.; Rogliani, P.; Leonardis, F.; Campana, A.; Carsetti, R.; et al. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA 2020, 96, 610–614. [Google Scholar] [CrossRef]
- Poulton, K.; Wright, P.; Hughes, P.; Savic, S.; Smith, M.W.; Guiver, M.; Morton, M.; Van Dellen, D.; Tholouli, E.; Wynn, R.; et al. A role for human leucocyte antigens in the susceptibility to SARS-Cov-2 infection observed in transplant patients. Int. J. Immunogenet. 2020, 47, 324–328. [Google Scholar] [CrossRef]
- Langton, D.J.; Bourke, S.C.; Lie, B.A.; Reiff, G.; Natu, S.; Darlay, R.; Burn, J.; Echevarria, C. The influence of HLA genotype on the severity of COVID-19 infection. HLA 2021, 98, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Anzurez, A.; Naka, I.; Miki, S.; Nakayama-Hosoya, K.; Isshiki, M.; Watanabe, Y.; Nakamura-Hoshi, M.; Seki, S.; Matsumura, T.; Takano, T.; et al. Association of HLA-DRB1 *09:01 with severe COVID-19. HLA 2021, 98, 37–42. [Google Scholar] [CrossRef]
- Amoroso, A.; Magistroni, P.; Vespasiano, F.; Bella, A.; Bellino, S.; Puoti, F.; Alizzi, S.; Vaisitti, T.; Boros, S.; Grossi, P.A.; et al. HLA and AB0 Polymorphisms May Influence SARS-CoV-2 Infection and COVID-19 Severity. Transplantation 2021, 105, 193–200. [Google Scholar] [CrossRef]
- Savage, P.A.; Klawon, D.E.; Miller, C.H. Regulatory T Cell Development. Annu. Rev. Immunol. 2020, 38, 421–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, D.C.; Zadoorian, A.; Wilson, L.O.W.; Thorne, N.P. Melbourne Genomics Health Alliance Evaluation of computational programs to predict HLA genotypes from genomic sequencing data. Brief. Bioinform. 2016, 19, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Yao, M.; Gong, Y.; Song, Y.; Chen, Y.; Ye, Y.; Liu, X.; Li, F.; Dong, H.; Meng, R.; et al. Benchmarking the Human Leukocyte Antigen Typing Performance of Three Assays and Seven Next-Generation Sequencing-Based Algorithms. Front. Immunol. 2021, 12, 652258. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Duffy, B.F.; Weimer, E.T.; Montgomery, M.C.; Jennemann, J.-E.; Hill, R.; Phelan, D.; Lay, L.; Parikh, B.A. Performance of a multiplexed amplicon-based next-generation sequencing assay for HLA typing. PLoS ONE 2020, 15, e0232050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cereb, N.; Kim, H.R.; Ryu, J.; Yang, S.Y. Advances in DNA sequencing technologies for high resolution HLA typing. Hum. Immunol. 2015, 76, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Duke, J.L.; Lind, C.; Mackiewicz, K.; Ferriola, D.; Papazoglou, A.; Gasiewski, A.; Heron, S.; Huynh, A.; McLaughlin, L.; Rogers, M.; et al. Determining performance characteristics of an NGS-based HLA typing method for clinical applications. HLA 2016, 87, 141–152. [Google Scholar] [CrossRef]
- Balz, V.; Fischer, J.; Krause, S.; Enczmann, J. A novel HLA-B variant, HLA-B*14:02:16, discovered by amplicon-based next-generation sequencing. HLA 2018, 92, 323–324. [Google Scholar] [CrossRef]
- Balz, V.; Krause, S.; Fischer, J.; Enczmann, J. More than 150 novel variants of HLA class I genes detected in German Stem Cell Donor Registry and UCLA International Cell Exchange samples. HLA 2018, 91, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Cargou, M.; Ralazamahaleo, M.; Blouin, L.; Top, I.; Elsermans, V.; Andreani, M.; Guidicelli, G.; Visentin, J. Evaluation of the AllType kit for HLA typing using the Ion Torrent S5 XL platform. HLA 2019, 95, 30–39. [Google Scholar] [CrossRef]
- Gutiérrez-Bautista, J.F.; Rodriguez-Nicolas, A.; Rosales-Castillo, A.; López-Ruz, M.; Martín-Casares, A.M.; Fernández-Rubiales, A.; Anderson, P.; Garrido, F.; Ruiz-Cabello, F.; López-Nevot, M. Study of HLA-A, -B, -C, -DRB1 and -DQB1 polymorphisms in COVID-19 patients. J. Microbiol. Immunol. Infect. 2021, 55, 421–427. [Google Scholar] [CrossRef]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Kanai, M.; Choi, W.; Li, X.; Sakaue, S.; Yamamoto, K.; Ogawa, K.; Gutierrez-Arcelus, M.; Gregersen, P.K.; Stuart, P.E.; et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet. 2021, 53, 1504–1516. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Shen, J.; Cox, C.J.; Wakefield, J.C.; Ehm, M.G.; Nelson, M.R.; Weir, B.S. HIBAG—HLA genotype imputation with attribute bagging. Pharm. J. 2013, 14, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitnis, N.; Clark, P.M.; Kamoun, M.; Stolle, C.; Johnson, F.B.; Monos, D.S. An Expanded Role for HLA Genes: HLA-B Encodes a microRNA that Regulates IgA and Other Immune Response Transcripts. Front. Immunol. 2017, 8, 583–594. [Google Scholar] [CrossRef]
- Colucci, M.; De Santis, E.; Totti, B.; Miroballo, M.; Tamiro, F.; Rossi, G.; Piepoli, A.; De Vincentis, G.; Greco, A.; Mangia, A.; et al. Associations between Allelic Variants of the Human IgH 3′ Regulatory Region 1 and the Immune Response to BNT162b2 mRNA Vaccine. Vaccines 2021, 9, 1207. [Google Scholar] [CrossRef]
- Čiučiulkaitė, I.; Möhlendick, B.; Thümmler, L.; Fisenkci, N.; Elsner, C.; Dittmer, U.; Siffert, W.; Lindemann, M. GNB3 c.825c>T polymorphism influences T-cell but not antibody response following vaccination with the mRNA-1273 vaccine. Front. Genet. 2022, 13, 932043. [Google Scholar] [CrossRef]
- Gemmati, D.; Longo, G.; Gallo, I.; Silva, J.A.; Secchiero, P.; Zauli, G.; Hanau, S.; Passaro, A.; Pellegatti, P.; Pizzicotti, S.; et al. Host genetics impact on SARS-CoV-2 vaccine-induced immunoglobulin levels and dynamics: The role of TP53, ABO, APOE, ACE2, HLA-A, and CRP genes. Front. Genet. 2022, 13, 1028081. [Google Scholar] [CrossRef]
- Kousathanas, A.; Pairo-Castineira, E.; Rawlik, K.; Stuckey, A.; Odhams, C.A.; Walker, S.; Russell, C.D.; Malinauskas, T.; Wu, Y.; Millar, J.; et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 2022, 607, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Fallerini, C.; Daga, S.; Mantovani, S.; Benetti, E.; Picchiotti, N.; Francisci, D.; Paciosi, F.; Schiaroli, E.; Baldassarri, M.; Fava, F.; et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: Findings from a nested case-control study. eLife 2021, 10, e67569. [Google Scholar] [CrossRef] [PubMed]
- Butler-Laporte, G.; Povysil, G.; Kosmicki, J.A.; Cirulli, E.T.; Drivas, T.; Furini, S.; Saad, C.; Schmidt, A.; Olszewski, P.; Korotko, U.; et al. Exome-wide association study to identify rare variants influencing COVID-19 outcomes: Results from the Host Genetics Initiative. PLoS Genet. 2022, 18, e1010367. [Google Scholar] [CrossRef]
- Arora, P.; Sidarovich, A.; Krüger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.-S.; Winkler, M.S.; Schulz, S.; Jäck, H.-M.; et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021, 37, 109825. [Google Scholar] [CrossRef]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; De Oliveira, T.; Vermeulen, M.; Van der Berg, K.; et al. SARS-CoV-2 501Y.V2 Escapes Neutralization by South African COVID-19 Donor Plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef]
- Emmelot, M.E.; Vos, M.; Boer, M.C.; Rots, N.Y.; van Els, C.A.C.M.; Kaaijk, P. SARS-CoV-2 Omicron BA.4/BA.5 Mutations in Spike Leading to T Cell Escape in Recently Vaccinated Individuals. Viruses 2022, 15, 101. [Google Scholar] [CrossRef]
- Xiao, C.; Mao, L.; Wang, Z.; Gao, L.; Zhu, G.; Su, J.; Chen, X.; Yuan, J.; Hu, Y.; Yin, Z.; et al. SARS-CoV-2 variant B.1.1.7 caused HLA-A2+ CD8+ T cell epitope mutations for impaired cellular immune response. iScience 2022, 25, 103934. [Google Scholar] [CrossRef]
- Nersisyan, S.; Zhiyanov, A.; Shkurnikov, M.; Tonevitsky, A. T-CoV: A comprehensive portal of HLA-peptide interactions affected by SARS-CoV-2 mutations. Nucleic Acids Res. 2021, 50, D883–D887. [Google Scholar] [CrossRef]
- Nersisyan, S.; Zhiyanov, A.; Zakharova, M.; Ishina, I.; Kurbatskaia, I.; Mamedov, A.; Galatenko, A.; Shkurnikov, M.; Gabibov, A.; Tonevitsky, A. Alterations in SARS-CoV-2 Omicron and Delta peptides presentation by HLA molecules. PeerJ 2022, 10, e13354. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-C.; Nersisyan, S.; Wu, C.-J.; Chang, C.-M.; Tonevitsky, A.; Guo, C.-L.; Chang, W.-C. On the peptide binding affinity changes in population-specific HLA repertoires to the SARS-CoV-2 variants Delta and Omicron. J. Autoimmun. 2022, 133, 102952. [Google Scholar] [CrossRef] [PubMed]
- Taher, J.; Mighton, C.; Chowdhary, S.; Casalino, S.; Frangione, E.; Arnoldo, S.; Bearss, E.; Binnie, A.; Bombard, Y.; Borgundvaag, B.; et al. Implementation of serological and molecular tools to inform COVID-19 patient management: Protocol for the GENCOV prospective cohort study. BMJ Open 2021, 11, e052842. [Google Scholar] [CrossRef] [PubMed]
| Study Cohort Characteristics | Antibody Response | References | |||
|---|---|---|---|---|---|
| Infection Status | Vaccination | Number of Individuals | Date Antibody Response Assessed | Antibody Response Assessed | |
| No prior infection | BNT162b2 | 100 | 7 and 39 days after second vaccine dose | Anti-RBD IgG | [60] |
| With or without prior infection | ChAdOx1 or ChAdOx1+ BNT162b2 or mRNA-1273 or NVX-CoV2373 | 1076 or 1677 | 28 days after first vaccine dose or 28 to 184 days after second vaccine dose | Anti-RBD and/or anti-S IgG | [61] |
| With or without prior infection | BNT162b2 | 420 | Unknown | Anti-RBD IgG | [62] |
| No prior infection | mRNA1273 | 87 | 30 days after second vaccine dose | Anti-S IgG | [63] |
| No prior infection | BNT162b2 | 87 | 144 days after second vaccine dose | Anti-RBD IgG and Neutralizing Antibodies | [64] |
| No prior infection | BNT162b2 | 111 | 14+ days after second vaccine dose | Anti-S IgG | [65] |
| Prior infection | None | 119 | >46 to >97 days after end of symptoms | Anti-RBD IgG, Anti-RBD IgA, Anti-NP total Ig, and Neutralizing antibodies | [66] |
| Prior Infection | None | 84 | 26 and 61 days after symptom onset | Neutralizing antibodies | [67] |
| No prior infection | BNT162b2 | 56 | 2 weeks to 4 months after second vaccine dose | Anti-RBD IgG | [68] |
| Prior infection | With or without BNT162b2 or ChAdOx1 | 49 | 12 months after PCR positivity | Anti-RBD IgG | [69] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolday, D.; Fung, C.Y.J.; Morgan, G.; Casalino, S.; Frangione, E.; Taher, J.; Lerner-Ellis, J.P. HLA Variation and SARS-CoV-2 Specific Antibody Response. Viruses 2023, 15, 906. https://doi.org/10.3390/v15040906
Wolday D, Fung CYJ, Morgan G, Casalino S, Frangione E, Taher J, Lerner-Ellis JP. HLA Variation and SARS-CoV-2 Specific Antibody Response. Viruses. 2023; 15(4):906. https://doi.org/10.3390/v15040906
Chicago/Turabian StyleWolday, Dawit, Chun Yiu Jordan Fung, Gregory Morgan, Selina Casalino, Erika Frangione, Jennifer Taher, and Jordan P. Lerner-Ellis. 2023. "HLA Variation and SARS-CoV-2 Specific Antibody Response" Viruses 15, no. 4: 906. https://doi.org/10.3390/v15040906
APA StyleWolday, D., Fung, C. Y. J., Morgan, G., Casalino, S., Frangione, E., Taher, J., & Lerner-Ellis, J. P. (2023). HLA Variation and SARS-CoV-2 Specific Antibody Response. Viruses, 15(4), 906. https://doi.org/10.3390/v15040906






