Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human and Animal Populations
2.2. SARS-CoV-2 RBD Peptides
2.3. Feline Cell Lines
2.4. Production and Partial Purification of FCoV2 Whole Virus
2.5. Transfection and Expression of Expi293F Cells with RBD Plasmids and Purification of RBD Proteins
2.6. FCoV-WV and SCoV2 RBD ELISAs with Overnight Serum Incubation
2.7. Stringent FCoV-WV and SCoV2 RBD ELISAs
2.8. Gel and Immunoblot Analyses
2.9. FCoV2 NAb Assay against FCoV2
2.10. RBD Blocking Assay against FCoV2
2.11. RBD Stimulation of PBMC from FCoV1-Infected Cats
2.12. Data Availability
3. Results
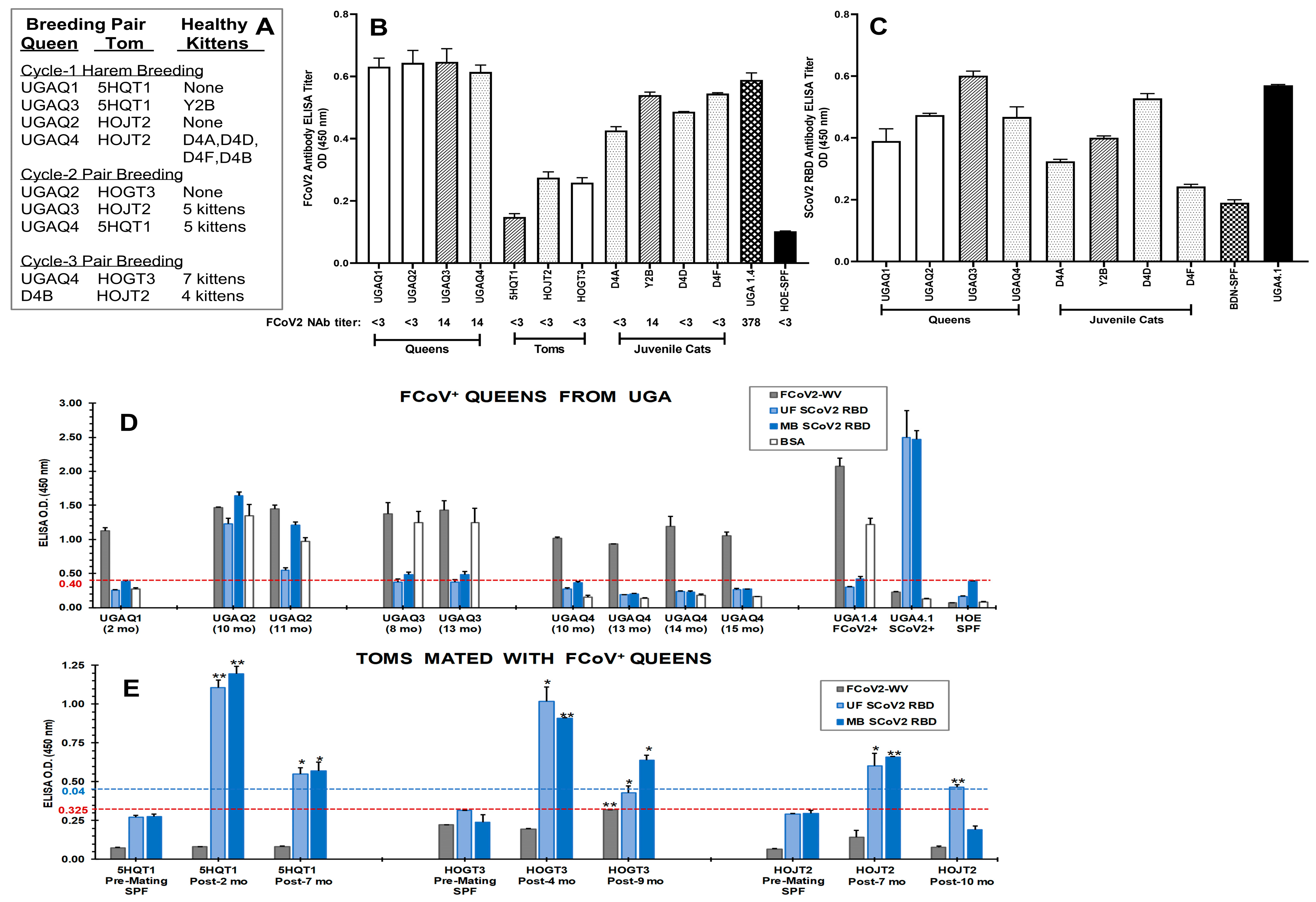
3.1. Initial Serological Studies in FCoV Naturally Infected Laboratory Cats
3.1.1. FCoV Whole-Virus (WV) ELISA and SCoV2 ELISA with Overnight Serum Incubation
3.1.2. Stringent FCoV-WV ELISA and SCoV2 RBD ELISA
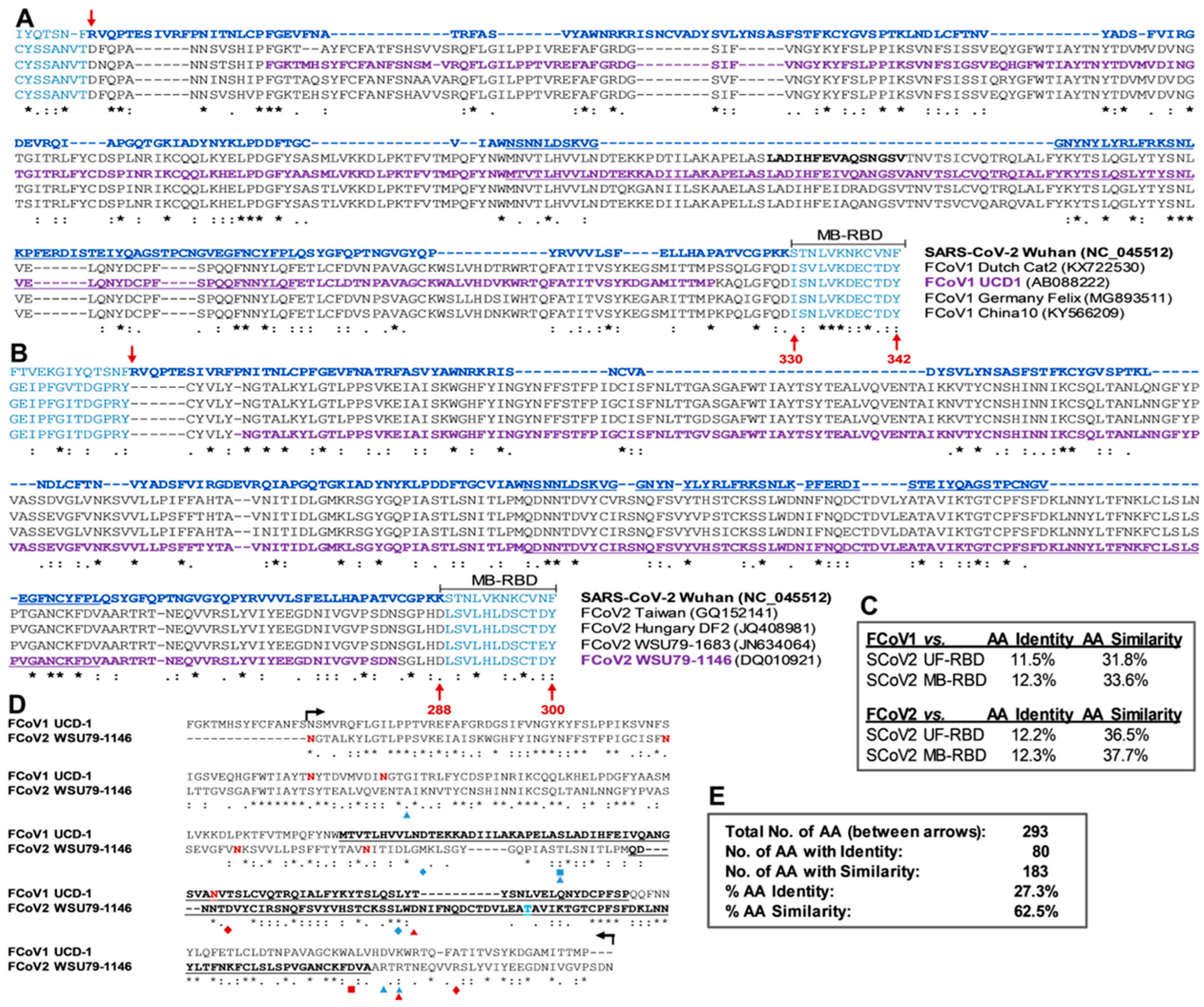
3.2. The Lack of S1 and RBD aa Sequence Identity/Similarity between SCoV2 and FCoVs
3.2.1. Sequence Analyses of SCoV2 and FCoV Structural Proteins
3.2.2. Sequence Analyses of SCoV2 and FCoV RBDs
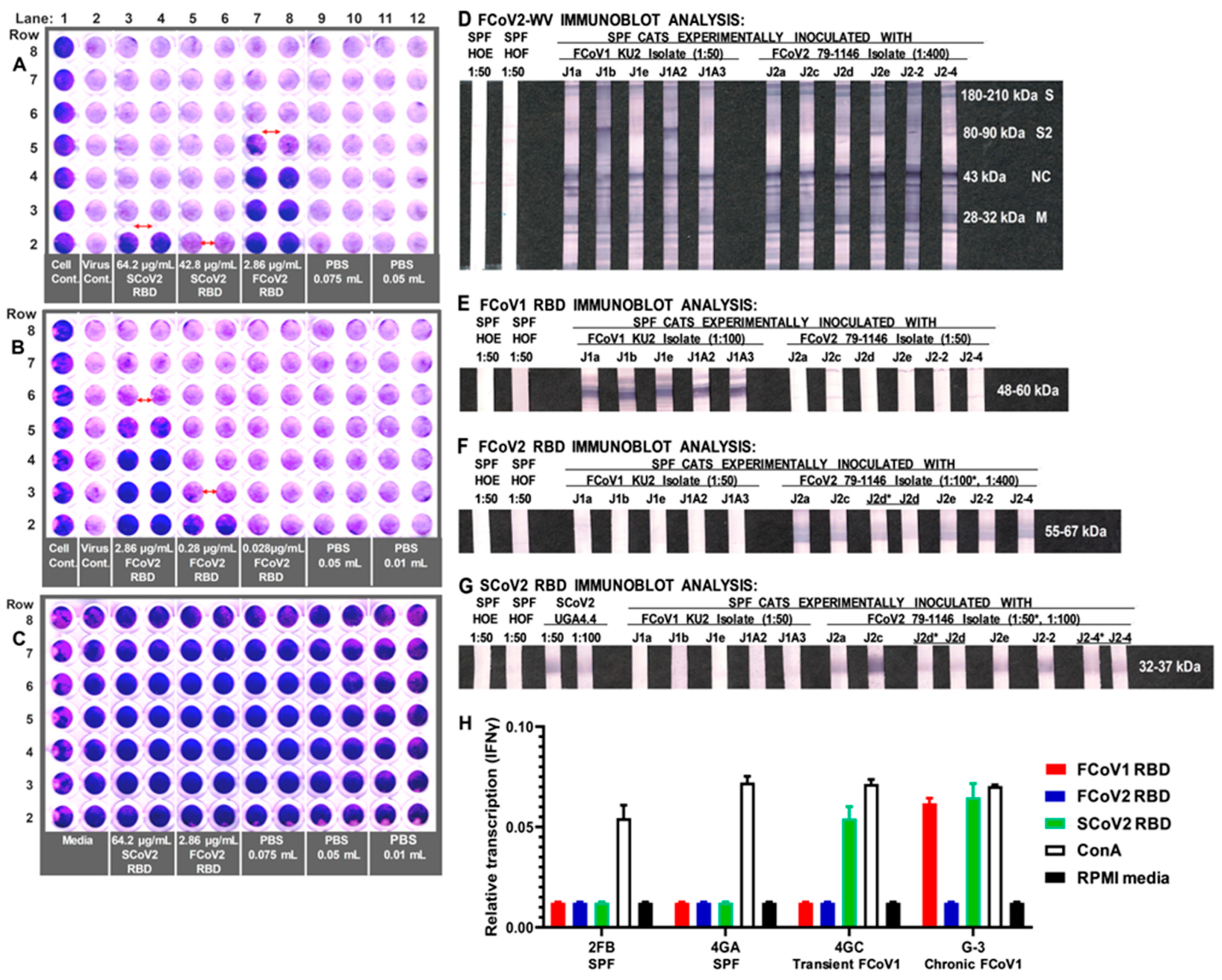
3.3. Immunoblot Analyses of Sera from Queens and Toms
3.4. Characterization of Abs to SCoV2 RBD
3.5. Determining the FCoV2 Infection Blocking Activity of FCoV2 RBD
3.6. Differential Reactivity to the FCoV1 and FCoV2 RBDs of the Plasma from the FCoV1 KU-2 and FCoV2 79-1146 Inoculated Laboratory Cats
3.7. The Role of Feline Genetics and Group Housing in Inducing Cross-Reactive SCoV2 RBD Abs
3.8. Interferon-Gamma (IFNγ) mRNA Production of PBMC from FCoV1-Infected Cats upon Stimulation with SCoV2, FCoV1, or FCoV2 RBD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Q.; Li, Y.; Huang, J.; Fu, N.; Song, X.; Sha, X.; Zhang, B. Prevalence and Molecular Characteristics of Feline Coronavirus in Southwest China from 2017 to 2020. J. Gen. Virol. 2021, 102, 001654. [Google Scholar] [CrossRef] [PubMed]
- Klein-Richers, U.; Hartmann, K.; Hofmann-Lehmann, R.; Unterer, S.; Bergmann, M.; Rieger, A.; Leutenegger, C.; Pantchev, N.; Balzer, J.; Felten, S. Prevalence of Feline Coronavirus Shedding in German Catteries and Associated Risk Factors. Viruses 2020, 12, 1000. [Google Scholar] [CrossRef] [PubMed]
- McKay, L.A.; Meachem, M.; Snead, E.; Brannen, T.; Mutlow, N.; Ruelle, L.; Davies, J.L.; van der Meer, F. Prevalence and Mutation Analysis of the Spike Protein in Feline Enteric Coronavirus and Feline Infectious Peritonitis Detected in Household and Shelter Cats in Western Canada. Can. J. Vet. Res. 2020, 84, 18–23. [Google Scholar] [PubMed]
- Horzinek, M.C.; Osterhaus, A.D. The Virology and Pathogenesis of Feline Infectious Peritonitis. Brief Review. Arch. Virol. 1979, 59, 1–15. [Google Scholar] [CrossRef]
- Vennema, H.; Poland, A.; Foley, J.; Pedersen, N.C. Feline Infectious Peritonitis Viruses Arise by Mutation from Endemic Feline Enteric Coronaviruses. Virology 1998, 243, 150–157. [Google Scholar] [CrossRef]
- Pedersen, N.C. An Update on Feline Infectious Peritonitis: Virology and Immunopathogenesis. Vet. J. 2014, 201, 123–132. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Diaz, A.; Damtie, D.; Xiu, L.; Toh, T.H.; Lee, J.S.; Saif, L.J.; Gray, G.C. Novel Canine Coronavirus Isolated from a Hospitalized Patient with Pneumonia in East Malaysia. Clin. Infect. Dis. 2022, 74, 446–454. [Google Scholar] [CrossRef]
- Silva, C.S.; Mullis, L.B.; Pereira, O.; Saif, L.J.; Vlasova, A.; Zhang, X.; Owens, R.J.; Paulson, D.; Taylor, D.; Haynes, L.M.; et al. Human Respiratory Coronaviruses Detected in Patients with Influenza-Like Illness in Arkansas, USA. Virol. Mycol. 2014, 2014 (Suppl. 2), 4. [Google Scholar]
- Vlasova, A.N.; Toh, T.H.; Lee, J.S.; Poovorawan, Y.; Davis, P.; Azevedo, M.S.P.; Lednicky, J.A.; Saif, L.J.; Gray, G.C. Animal Alphacoronaviruses Found in Human Patients with Acute Respiratory Illness in Different Countries. Emerg. Microbes Infect. 2022, 11, 699–702. [Google Scholar] [CrossRef]
- Pagani, G.; Lai, A.; Bergna, A.; Rizzo, A.; Stranieri, A.; Giordano, A.; Paltrinieri, S.; Lelli, D.; Decaro, N.; Rusconi, S.; et al. Human-to-Cat SARS-CoV-2 Transmission: Case Report and Full-Genome Sequencing from an Infected Pet and Its Owner in Northern Italy. Pathogens 2021, 10, 252. [Google Scholar] [CrossRef]
- World Organization for Animal Health. Global Cases of SARS-CoV-2 in Animals. (Note This Website Provides Information about Each Animal Case with Some Clinical History). (Report ID FUR_154029 Describes Hamster-to-Human Transmission of SCoV2 Delta Variant). Available online: https://www.oie.int/en/what-we-offer/emergency-and-resilience/covid-19/#ui-id-3 (accessed on 14 June 2022).
- USDA. Confirmed Cases of SARS-CoV-2. In Animals in the United States; Last Updated 23 March 2022; USDA: Washington, DC, USA, 2022. Available online: https://www.aphis.usda.gov/aphis/ourfocus/onehealth/one-health-sarscov2-in-animals (accessed on 1 August 2022).
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of Ferrets, Cats, Dogs, and Other Domesticated Animals to SARS-Coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; Vande Woude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental Infection of Domestic Dogs and Cats with SARS-CoV-2: Pathogenesis, Transmission, and Response to Reexposure in Cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 Infection, Disease and Transmission in Domestic Cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of Dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Zhang, X.; Hasoksuz, M.; Nagesha, H.S.; Haynes, L.M.; Fang, Y.; Lu, S.; Saif, L.J. Two-Way Antigenic Cross-Reactivity Between Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Group 1 Animal CoVs Is Mediated Through an Antigenic Site in the N-Terminal Region of the SARS-CoV Nucleoprotein. J. Virol. 2007, 81, 13365–13377. [Google Scholar] [CrossRef]
- Zhao, S.; Li, W.; Schuurman, N.; van Kuppeveld, F.; Bosch, B.J.; Egberink, H. Serological Screening for Coronavirus Infections in Cats. Viruses 2019, 11, 743. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Skowronski, D.M.; Astell, C.; Brunham, R.C.; Low, D.E.; Petric, M.; Roper, R.L.; Talbot, P.J.; Tam, T.; Babiuk, L. Severe Acute Respiratory Syndrome (SARS): A Year in Review. Annu. Rev. Med. 2005, 56, 357–381. [Google Scholar] [CrossRef]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef]
- Martina, B.E.E.; Haagmans, B.L.; Kuiken, T.; Fouchier, R.A.; Rimmelzwaan, G.F.; Van Amerongen, G.; Peiris, J.S.; Lim, W.; Osterhaus, A.D. Virology: SARS Virus Infection of Cats and Ferrets. Nature 2003, 425, 915. [Google Scholar] [CrossRef]
- Stout, A.E.; André, N.M.; Jaimes, J.A.; Millet, J.K.; Whittaker, G.R. Coronaviruses in Cats and Other Companion Animals: Where Does SARS-CoV-2/COVID-19 Fit? Vet. Microbiol. 2020, 247, 108777. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Hohdatsu, T.; Izumiya, Y.; Yokoyama, Y.; Kida, K.; Koyama, H. Differences in Virus Receptor for Type I and Type II Feline Infectious Peritonitis Virus. Arch. Virol. 1998, 143, 839–850. [Google Scholar] [CrossRef]
- Dye, C.; Temperton, N.; Siddell, S.G. Type I Feline Coronavirus Spike Glycoprotein Fails to Recognize Aminopeptidase N as a Functional Receptor on Feline Cell Lines. J. Gen. Virol. 2007, 88, 1753–1760. [Google Scholar] [CrossRef]
- Tusell, S.M.; Schittone, S.A.; Holmes, K.V. Mutational Analysis of Aminopeptidase N, a Receptor for Several Group 1 Coronaviruses, Identifies Key Determinants of Viral Host Range. J. Virol. 2007, 81, 1261–1273. [Google Scholar] [CrossRef]
- Kolb, A.F.; Hegyi, A.; Siddell, S.G. Identification of Residues Critical for the Human Coronavirus 229E Receptor Function of Human Aminopeptidase N. J. Gen. Virol. 1997, 78, 2795–2802. [Google Scholar] [CrossRef]
- Jaimes, J.A.; Millet, J.K.; Stout, A.E.; André, N.M.; Whittaker, G.R. A Tale of Two Viruses: The Distinct Spike Glycoproteins of Feline Coronaviruses. Viruses 2020, 12, 83. [Google Scholar] [CrossRef]
- An, D.J.; Jeoung, H.Y.; Jeong, W.; Park, J.Y.; Lee, M.H.; Park, B.K. Prevalence of Korean Cats with Natural Feline Coronavirus Infections. Virol. J. 2011, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Terada, Y.; Matsui, N.; Noguchi, K.; Kuwata, R.; Shimoda, H.; Soma, T.; Mochizuki, M.; Maeda, K. Emergence of Pathogenic Coronaviruses in Cats by Homologous Recombination Between Feline and Canine Coronaviruses. PLoS ONE 2014, 9, e106534. [Google Scholar] [CrossRef]
- Wu, L.; Chen, Q.; Liu, K.; Wang, J.; Han, P.; Zhang, Y.; Hu, Y.; Meng, Y.; Pan, X.; Qiao, C.; et al. Broad Host Range of SARS-CoV-2 and the Molecular Basis for SARS-CoV-2 Binding to Cat ACE2. Cell Discov. 2020, 6, 68. [Google Scholar] [CrossRef]
- Conceicao, C.; Thakur, N.; Human, S.; Kelly, J.T.; Logan, L.; Bialy, D.; Bhat, S.; Stevenson-Leggett, P.; Zagrajek, A.K.; Hollinghurst, P.; et al. The SARS-CoV-2 Spike Protein Has a Broad Tropism for Mammalian ACE2 Proteins. PLoS Biol. 2020, 18, e3001016. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade- Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia After Visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203.e3. [Google Scholar] [CrossRef] [PubMed]
- Regan, A.D.; Millet, J.K.; Tse, L.P.; Chillag, Z.; Rinaldi, V.D.; Licitra, B.N.; Dubovi, E.J.; Town, C.D.; Whittaker, G.R. Characterization of a Recombinant Canine Coronavirus with a Distinct Receptor-Binding (S1) Domain. Virology 2012, 430, 90–99. [Google Scholar] [CrossRef]
- Li, C.; Li, W.; Lucio de Esesarte, E.; Guo, H.; van den Elzen, P.; Aarts, E.; van den Born, E.; Rottier, P.J.M.; Bosch, B.J. Cell Attachment Domains of the Porcine Epidemic Diarrhea Virus Spike Protein Are Key Targets of Neutralizing Antibodies. J. Virol. 2017, 91, e00273-17. [Google Scholar] [CrossRef]
- Reguera, J.; Ordoño, D.; Santiago, C.; Enjuanes, L.; Casasnovas, J.M. Antigenic Modules in the N-Terminal S1 Region of the Transmissible Gastroenteritis Virus Spike Protein. J. Gen. Virol. 2011, 92, 1117–1126. [Google Scholar] [CrossRef]
- Shirato, K.; Maejima, M.; Islam, M.T.; Miyazaki, A.; Kawase, M.; Matsuyama, S.; Taguchi, F. Porcine Aminopeptidase N Is Not a Cellular Receptor of Porcine Epidemic Diarrhoea Virus, but Promotes Its Infectivity via Aminopeptidase Activity. J. Gen. Virol. 2016, 97, 2528–2539. [Google Scholar] [CrossRef]
- Kida, K.; Hohdatsu, T.; Fujii, K.; Koyama, H. Selection of Antigenic Variants of the S Glycoprotein of Feline Infectious Peritonitis Virus and Analysis of Antigenic Sites Involved in Neutralization. J. Vet. Med. Sci. 1999, 61, 935–938. [Google Scholar] [CrossRef]
- Corapi, W.V.; Darteil, R.J.; Audonnet, J.C.; Chappuis, G.E. Localization of Antigenic Sites of the S Glycoprotein of Feline Infectious Peritonitis Virus Involved in Neutralization and Antibody-Dependent Enhancement. J. Virol. 1995, 69, 2858–2862. [Google Scholar] [CrossRef]
- Dye, C.; Helps, C.R.; Siddell, S.G. Evaluation of Real-Time RT-PCR for the Quantification of FCoV Shedding in the Faeces of Domestic Cats. J. Feline Med. Surg. 2008, 10, 167–174. [Google Scholar] [CrossRef]
- Nanishi, E.; Borriello, F.; O’Meara, T.R.; McGrath, M.E.; Saito, Y.; Haupt, R.E.; Seo, H.S.; van Haren, S.D.; Cavazzoni, C.B.; Brook, B.; et al. An Aluminium Hydroxide: CpG Adjuvant Enhances Protection Elicited by a SARS-CoV-2 Receptor Binding Domain Vaccine in Aged Mice. Sci. Transl. Med. 2022, 14, eabj5305. [Google Scholar] [CrossRef]
- Yates, J.L.; Ehrbar, D.J.; Hunt, D.T.; Girardin, R.C.; Dupuis, A.P., 2nd; Payne, A.F.; Sowizral, M.; Varney, S.; Kulas, K.E.; Demarest, V.L.; et al. Serological Analysis Reveals an Imbalanced IgG Subclass Composition Associated with COVID-19 Disease Severity. Reprod. Med. 2021, 2, 100329. [Google Scholar] [CrossRef] [PubMed]
- AP News. Florida Has Two Confirmed Cases of New Coronavirus|AP News. Available online: https://apnews.com/article/travel-public-health-health-virus-outbreak-tampa-bd1bc203ce72aecc44774b64f41e77e6 (accessed on 15 January 2023).
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The Spike Glycoprotein of the New Coronavirus 2019-nCoV Contains a Furin-Like Cleavage Site Absent in CoV of the Same Clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Hosts Hosts Cell Proteases: Critical Determinants of Coronavirus Tropism and Pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Aillo, S.E.; Moses, M.A. Feline Infectious Peritonitis: Management of Multi-cat Households. In The Merck Veterinary Manual, 11th ed.; Chapter: Generalized Conditions; Merck & Co., Inc.: Kenilworth, NJ, USA, 2016; p. 788. [Google Scholar]
- Follis, K.E.; York, J.; Nunberg, J.H. Furin Cleavage of the SARS Coronavirus Spike Glycoprotein Enhances Cell-Cell Fusion but Does Not Affect Virion Entry. Virology 2006, 350, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Calarese, D.A.; Lee, H.K.; Huang, C.Y.; Best, M.D.; Astronomo, R.D.; Stanfield, R.L.; Katinger, H.; Burton, D.R.; Wong, C.H.; Wilson, I.A. Dissection of the Carbohydrate Specificity of the Broadly Neutralizing Anti-HIV-1 Antibody 2G12. Proc. Natl. Acad. Sci. USA 2005, 102, 13372–13377. [Google Scholar] [CrossRef]
- Aranyos, A.M.; Roff, S.R.; Pu, R.; Owen, J.L.; Coleman, J.K.; Yamamoto, J.K. An Initial Examination of the Potential Role of T-Cell Immunity in Protection Against Feline Immunodeficiency Virus (FIV) Infection. Vaccine 2016, 34, 1480–1488. [Google Scholar] [CrossRef]
- Sahay, B.; Aranyos, A.M.; Mishra, M.; McAvoy, A.C.; Martin, M.M.; Pu, R.; Shiomitsu, S.; Shiomitsu, K.; Dark, M.J.; Sanou, M.P.; et al. Immunogenicity and Efficacy of a Novel Multi-antigenic Peptide Vaccine Based on Cross-Reactivity between Feline and Human Immunodeficiency Viruses. Viruses 2019, 11, 136. [Google Scholar] [CrossRef]
- Sakurai, A.; Sasaki, T.; Kato, S.; Hayashi, M.; Tsuzuki, S.I.; Ishihara, T.; Iwata, M.; Morise, Z.; Doi, Y. Natural History of Asymptomatic SARS-CoV-2 Infection. N. Engl. J. Med. 2020, 383, 885–886. [Google Scholar] [CrossRef] [PubMed]
- Meyerowitz, E.A.; Richterman, A.; Bogoch, I.I.; Low, N.; Cevik, M. Towards an Accurate and Systematic Characterisation of Persistently Asymptomatic Infection with SARS-CoV-2. Lancet Infect. Dis. 2021, 21, e163–e169. [Google Scholar] [CrossRef]
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.J.; Bosch, B.J.; et al. Human Coronaviruses OC43 and HKU1 Bind to 9-O-Acetylated Sialic Acids via a Conserved Receptor-Binding Site in Spike Protein Domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Hancock, T.J.; Hickman, P.; Kazerooni, N.; Kennedy, M.; Kania, S.A.; Dennis, M.; Szafranski, N.; Gerhold, R.; Su, C.; Masi, T.; et al. Possible Cross-Reactivity of Feline and White-Tailed Deer Antibodies Against the SARS-CoV-2 Receptor Binding Domain. J. Virol. 2022, 96, e0025022. [Google Scholar] [CrossRef] [PubMed]
- Terpe, K. Overview of Tag Protein Fusions: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Takara. Obtain Highest Purity with Cobalt Resin. Available online: https://www.takarabio.com/learning-centers/protein-research/his-tag-purification/tech-note-cobalt-resin (accessed on 14 January 2023).
- da Silva Junior, H.C. Transient Gene expression in Human Expi293 Cells. In Insoluble Proteins: Methods and Protocols, 2nd ed.; Fruitos, E.G., Giralt, A.A., Eds.; Humana: New York, NY, USA, 2022; Volume 2406, pp. 319–325. [Google Scholar]
- Anton Paar. Dimerizaation of Bovine Serum Albumin as Evidenced by Particle Size and Molecular Mass Measurement. Available online: https://www.anton-paar.com/corp-en/services-support/document-finder/application-reports/dimerization-of-bovine-serum-albumin-as-evidenced-by-particle-size-and-molecular-mass-measurement/ (accessed on 15 January 2023).
- Molodenskiy, D.; Shirshin, E.; Tikhonova, T.; Gruzinov, A.; Peters, G.; Spinozzi, F. Thermally Induced Conformational Changes and Protein-Protein Interactions of Bovine Serum Albumin in Aqueous Solution Under Different pH and Ionic Strengths as Revealed by SAXS Measurements. Phys. Chem. Chem. Phys. 2017, 19, 17143–17155. [Google Scholar] [CrossRef]
- Shin, H.J.; Ku, K.B.; Kim, H.S.; Moon, H.W.; Jeong, G.U.; Hwang, I.; Yoon, G.Y.; Lee, S.; Lee, S.; Ahn, D.G.; et al. Receptor-Binding Domain of SARS-CoV-2 Spike Protein Efficiently Inhibits SARS-CoV-2 Infection and Attachment to Mouse Lung. Int. J. Biol. Sci. 2021, 17, 3786–3794. [Google Scholar] [CrossRef]
- Roff, S.R.; Noon-Song, E.N.; Yamamoto, J.K. The Significance of Interferon-γ in HIV-1 Pathogenesis, Therapy, and Prophylaxis. Front. Immunol. 2014, 4, 498. [Google Scholar] [CrossRef]
- Takano, T.; Kawakami, C.; Yamada, S.; Satoh, R.; Hohdatsu, T. Antibody-Dependent Enhancement Occurs Upon Re-Infection with the Identical Serotype Virus in Feline Infectious Peritonitis Virus Infection. J. Vet. Med. Sci. 2008, 70, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Boyle, J.F.; Floyd, K. Infection Studies in Kittens, Using Feline Infectious Peritonitis Virus Propagated in Cell Culture. Am. J. Vet. Res. 1981, 42, 363–367. [Google Scholar] [PubMed]
- de Haan, C.A.; Haijema, B.J.; Schellen, P.; Wichgers Schreur, P.; te Lintelo, E.; Vennema, H.; Rottier, P.J. Cleavage of Group 1 Coronavirus Spike Proteins: How Furin Cleavage is Traded Off against Heparan Sulfate Binding upon Cell Culture Adaptation. J. Virol. 2008, 82, 6078–6083. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Güler, A.; et al. BNT162b Vaccines Protect Rhesus Macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef]
- Vanderheijden, N.; Stevaert, A.; Xie, J.; Ren, X.; Barbezange, C.; Noppen, S.; Desombere, I.; Verhasselt, B.; Geldhof, P.; Vereecke, N.; et al. Functional Analysis of Human and Feline Coronavirus Cross-Reactive Antibodies Directed Against the SARS-CoV-2 Fusion Peptide. Front. Immunol. 2021, 12, 790415. [Google Scholar] [CrossRef]
- Dacon, C.; Tucker, C.; Peng, L.; Lee, C.D.; Lin, T.H.; Yuan, M.; Cong, Y.; Wang, L.; Purser, L.; Williams, J.K.; et al. Broadly Neutralizing Antibodies Target the Coronavirus Fusion Peptide. Science 2022, 377, 728–735. [Google Scholar] [CrossRef]
- Pinto, D.; Sauer, M.M.; Czudnochowski, N.; Low, J.S.; Tortorici, M.A.; Housley, M.P.; Noack, J.; Walls, A.C.; Bowen, J.E.; Guarino, B.; et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science 2021, 373, 1109–1116. [Google Scholar] [CrossRef]
- He, C.; Yang, J.; Hong, W.; Chen, Z.; Peng, D.; Lei, H.; Alu, A.; He, X.; Bi, Z.; Jiang, X.; et al. A Self-Assembled Trimeric Protein Vaccine Induces Protective Immunity Against Omicron Variant. Nat. Commun. 2022, 13, 5459. [Google Scholar] [CrossRef]
- Dela Cruz, C.S.; Chamberlain, J.W.; MacDonald, K.S.; Barber, B.H. Xenogeneic and Allogeneic Anti-MHC Immune Responses Induced by Plasmid DNA Immunization. Vaccine 1999, 17, 2479–2492. [Google Scholar] [CrossRef]
- Desrosiers, R.C.; Wyand, M.S.; Kodama, T.; Ringler, D.J.; Arthur, L.O.; Sehgal, P.K.; Letvin, N.L.; King, N.W.; Daniel, M.D. Vaccine Protection Against Simian Immunodeficiency Virus Infection. Proc. Natl. Acad. Sci. USA 1989, 86, 6353–6357. [Google Scholar] [CrossRef]
- Murphey-Corb, M.; Martin, L.N.; Davison-Fairburn, B.; Montelaro, R.C.; Miller, M.; West, M.; Ohkawa, S.; Baskin, G.B.; Zhang, J.Y.; Putney, S.D.; et al. A Formalin-Inactivated Whole SIV Vaccine Confers Protection in Macaques. Science 1989, 246, 1293–1297. [Google Scholar] [CrossRef]
- Edwards, K.M.; Orenstein, W.A. COVID-19: Vaccines-UpToDate. Available online: https://www.uptodate.com/contents/covid-19-vaccines/print (accessed on 15 January 2023).
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and Transmission of SARS-CoV-2 in Golden Hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Boudewijns, R.; Foo, C.S.; Seldeslachts, L.; Sanchez-Felipe, L.; Zhang, X.; Delang, L.; Maes, P.; Kaptein, S.J.F.; Weynand, B.; et al. Comparing Infectivity and Virulence of Emerging SARS-CoV-2 Variants in Syrian Hamsters. eBiomedicine 2021, 68, 103403. [Google Scholar] [CrossRef]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent Infections of Porcine Deltacoronavirus Among Haitian Children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Blohm, G.M.; Alam, M.M.; Iovine, N.M.; Salemi, M.; Mavian, C.; Morris, J.G. Isolation of a Novel Recombinant Canine Coronavirus from a Visitor to Haiti: Further Evidence of Transmission of Coronaviruses of Zoonotic Origin to Humans. Clin. Infect. Dis. 2022, 75, e1184–e1187. [Google Scholar] [CrossRef] [PubMed]
- Sila, T.; Sunghan, J.; Laochareonsuk, W.; Surasombatpattana, S.; Kongkamol, C.; Ingviya, T.; Siripaitoon, P.; Kositpantawong, N.; Kanchanasuwan, S.; Hortiwakul, T.; et al. Suspected Cat-to-Human Transmission of SARS-CoV-2, Thailand, July-September 2021. Emerg. Infect. Dis. 2022, 28, 1485–1488. [Google Scholar] [CrossRef] [PubMed]
- Díaz, L.A.; García-Salum, T.; Fuentes-López, E.; Reyes, D.; Ortiz, J.; Chahuan, J.; Levican, J.; Almonacid, L.I.; Valenzuela, G.H.; Serrano, E.; et al. High Prevalence of SARS-CoV-2 Detection and Prolonged Viral Shedding in Stools: A Systematic Review and Cohort Study. Gastroenterol. Hepatol. 2022, 45, 593–604. [Google Scholar] [CrossRef]
- Galanopoulos, M.; Karianakis, G.; Amorginos, K.; Doukatas, A.; Gkeros, F.; Tsoukalas, N.; Papanikolaou, I.; Viazis, N.; Liatsos, C. Laboratory Manifestations and Pathophysiological Aspects of Coronavirus Disease 2019 Pandemic: Focusing on the Digestive System. Eur. J. Gastroenterol. Hepatol. 2021, 33 (Suppl. 1), e59–e65. [Google Scholar] [CrossRef]
- Licitra, B.N.; Duhamel, G.E.; Whittaker, G.R. Canine Enteric Coronaviruses: Emerging Viral Pathogens with Distinct Recombinant Spike Proteins. Viruses 2014, 6, 3363–3376. [Google Scholar] [CrossRef]
- Wang, Y.T.; Su, B.L.; Hsieh, L.E.; Chueh, L.L. An Outbreak of Feline Infectious Peritonitis in a Taiwanese Shelter: Epidemiologic and Molecular Evidence for Horizontal Transmission of a Novel Type II feline Coronavirus. Vet. Res. 2013, 44, 57. [Google Scholar] [CrossRef] [PubMed]
- Yen, S.-J.; Chen, H.-W. Feline Coronaviruses Identified in Feline Effusions in Suspected Cases of Feline Infectious Peritonitis. Microorganisms 2021, 9, 1801. [Google Scholar] [CrossRef] [PubMed]
- Katayama, M.; Uemura, Y. Prognostic Prediction for Therapeutic Effects of Mutian on 324 Client-Owned Cats with Feline Infectious Peritonitis Based on Clinical Laboratory Indicators and Physical Signs. Vet. Sci. 2023, 10, 136. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, J.K.; Edison, L.K.; Rowe-Haas, D.K.; Takano, T.; Gilor, C.; Crews, C.D.; Tuanyok, A.; Arukha, A.P.; Shiomitsu, S.; Walden, H.D.S.; et al. Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development. Viruses 2023, 15, 914. https://doi.org/10.3390/v15040914
Yamamoto JK, Edison LK, Rowe-Haas DK, Takano T, Gilor C, Crews CD, Tuanyok A, Arukha AP, Shiomitsu S, Walden HDS, et al. Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development. Viruses. 2023; 15(4):914. https://doi.org/10.3390/v15040914
Chicago/Turabian StyleYamamoto, Janet K., Lekshmi K. Edison, Dawne K. Rowe-Haas, Tomomi Takano, Chen Gilor, Chiquitha D. Crews, Apichai Tuanyok, Ananta P. Arukha, Sayaka Shiomitsu, Heather D. S. Walden, and et al. 2023. "Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development" Viruses 15, no. 4: 914. https://doi.org/10.3390/v15040914
APA StyleYamamoto, J. K., Edison, L. K., Rowe-Haas, D. K., Takano, T., Gilor, C., Crews, C. D., Tuanyok, A., Arukha, A. P., Shiomitsu, S., Walden, H. D. S., Hohdatsu, T., Tompkins, S. M., Morris Jr., J. G., Sahay, B., & Kariyawasam, S. (2023). Both Feline Coronavirus Serotypes 1 and 2 Infected Domestic Cats Develop Cross-Reactive Antibodies to SARS-CoV-2 Receptor Binding Domain: Its Implication to Pan-CoV Vaccine Development. Viruses, 15(4), 914. https://doi.org/10.3390/v15040914









