Intrahost Genetic Diversity of Dengue Virus in Human Hosts and Mosquito Vectors under Natural Conditions Which Impact Replicative Fitness In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Population
2.3. RNA Extraction, Library Preparation, and Sequencing
2.4. Mapping and Single-Nucleotide Variant (SNV) Calling
2.5. Cells, Infectious Clone, and Antibody
2.6. Site-Directed Mutagenesis
2.7. Indirect Immunofluorescence Assays
2.8. Viral RNA Quantification
2.9. Replication Kinetics
2.10. Data Analysis and Software
3. Results
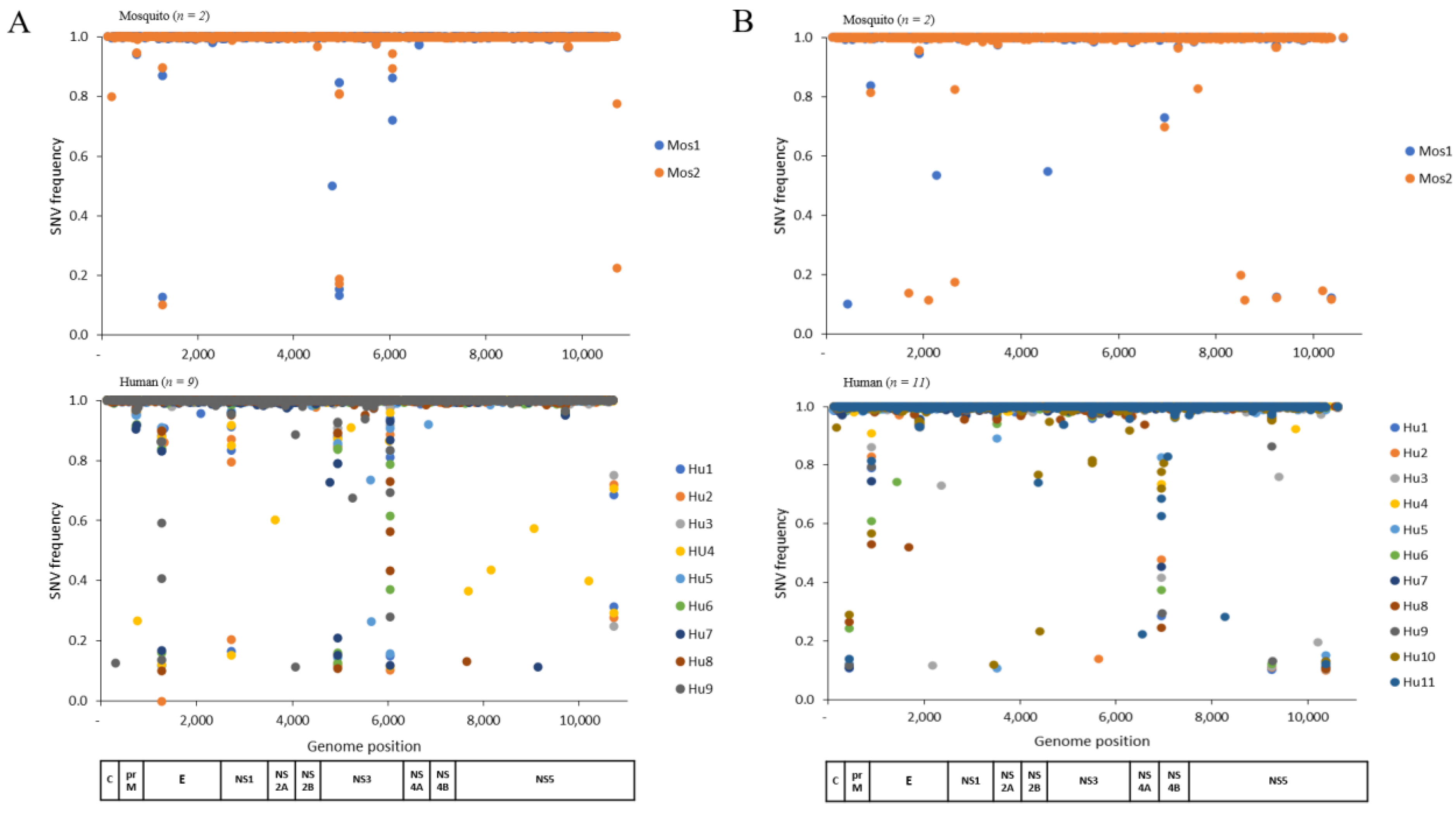
3.1. Intrahost Genetic Diversity of DENV-1 and -4 in Humans and Mosquitoes
3.2. Specific Nonsynonymous Substitution Is Acquired in Ae. aegypti Mosquito
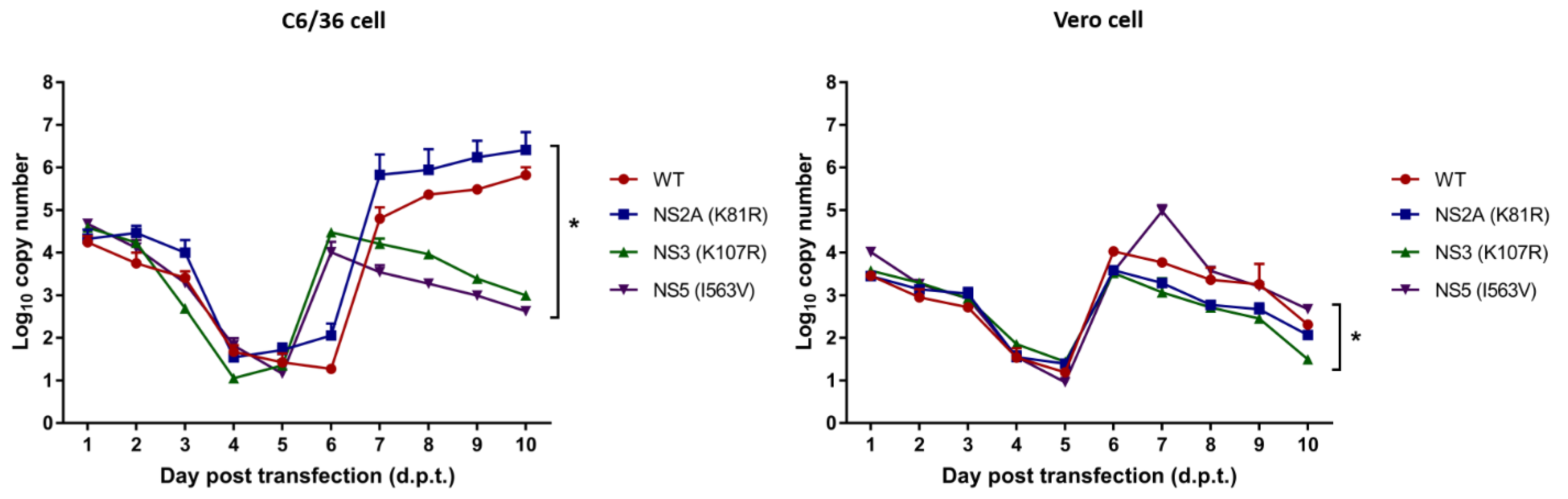
3.3. RNA Production from DENV-4 Infectious Clone
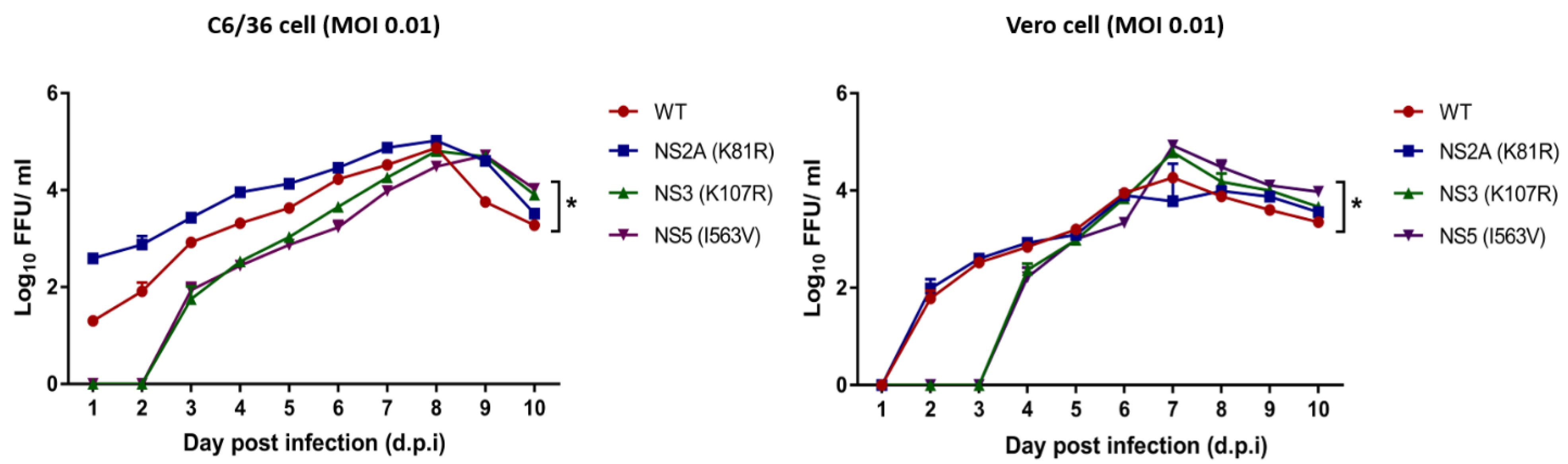
3.4. Replication Kinetics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.; Hay, S.; Bedi, N.; Bensenor, I.M.; Castañeda-Orjuela, C.; et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, M.U.; Sinka, M.E.; Duda, K.A.; Mylne, A.Q.; Shearer, F.M.; Barker, C.M.; Moore, C.G.; Carvalho, R.G.; Coelho, G.E.; Van Bortel, W.; et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife 2015, 4, e08347. [Google Scholar] [CrossRef]
- Low, J.G.H.; Ooi, E.E.; Vasudevan, S. Current Status of Dengue Therapeutics Research and Development. J. Infect. Dis. 2017, 215, S96–S102. [Google Scholar] [CrossRef] [PubMed]
- Prompetchara, E.; Ketloy, C.; Thomas, S.J.; Ruxrungtham, K. Dengue vaccine: Global development update. Asian Pac. J. Allergy Immunol. 2019, 10, 178–185. [Google Scholar] [CrossRef]
- Parameswaran, P.; Charlebois, P.; Tellez, Y.; Nunez, A.; Ryan, E.M.; Malboeuf, C.M.; Levin, J.Z.; Lennon, N.J.; Balmaseda, A.; Harris, E.; et al. Genome-Wide Patterns of Intrahuman Dengue Virus Diversity Reveal Associations with Viral Phylogenetic Clade and Interhost Diversity. J. Virol. 2012, 86, 8546–8558. [Google Scholar] [CrossRef] [PubMed]
- Rana, J.; Campos, J.L.S.; Poggianella, M.; Burrone, O.R. Dengue virus capsid anchor modulates the efficiency of polyprotein processing and assembly of viral particles. J. Gen. Virol. 2019, 100, 1663–1673. [Google Scholar] [CrossRef]
- Uno, N.; Ross, T.M. Dengue virus and the host innate immune response. Emerg. Microbes Infect. 2018, 7, 167. [Google Scholar] [CrossRef]
- El Sahili, A.; Lescar, J. Dengue Virus Non-Structural Protein 5. Viruses 2017, 9, 91. [Google Scholar] [CrossRef]
- Sim, S.; Hibberd, M.L. Genomic approaches for understanding dengue: Insights from the virus, vector, and host. Genome Biol. 2016, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, D.; Das, S.; Begum, F.; Mal, S.; Ray, U. The Mosquito Immune System and the Life of Dengue Virus: What We Know and Do Not Know. Pathogens 2019, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Sessions, O.M.; Wilm, A.; Kamaraj, U.S.; Choy, M.M.; Chow, A.; Chong, Y.; Ong, X.M.; Nagarajan, N.; Cook, A.; Ooi, E.E. Analysis of Dengue Virus Genetic Diversity during Human and Mosquito Infection Reveals Genetic Constraints. PLoS Negl. Trop. Dis. 2015, 9, e0004044. [Google Scholar] [CrossRef]
- Martinez, M.A.; Franco, S. Therapy Implications of Hepatitis C Virus Genetic Diversity. Viruses 2020, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Folkers, G.K. Toward an AIDS-Free Generation. JAMA 2012, 308, 343–344. [Google Scholar] [CrossRef] [PubMed]
- Paterson, J.C.; Miller, M.H.; Dillon, J. Update on the treatment of hepatitis C genotypes 2–6. Curr. Opin. Infect. Dis. 2014, 27, 540–544. [Google Scholar] [CrossRef]
- Galli, A.; Mens, H.; Gottwein, J.M.; Gerstoft, J.; Bukh, J. Antiviral Effect of Ribavirin against HCV Associated with Increased Frequency of G-to-A and C-to-U Transitions in Infectious Cell Culture Model. Sci. Rep. 2018, 8, 4619. [Google Scholar] [CrossRef]
- Sim, S.; Aw, P.P.K.; Wilm, A.; Teoh, G.; Hue, K.D.T.; Nguyen, N.; Nagarajan, N.; Simmons, C.P.; Hibberd, M.L. Tracking Dengue Virus Intra-host Genetic Diversity during Human-to-Mosquito Transmission. PLoS Negl. Trop. Dis. 2015, 9, e0004052. [Google Scholar] [CrossRef] [PubMed]
- Fustec, B.; Phanitchat, T.; Hoq, M.I.; Aromseree, S.; Pientong, C.; Thaewnongiew, K.; Ekalaksananan, T.; Bangs, M.J.; Corbel, V.; Alexander, N.; et al. Complex relationships between Aedes vectors, socio-economics and dengue transmission—Lessons learned from a case-control study in northeastern Thailand. PLoS Negl. Trop. Dis. 2020, 14, e0008703. [Google Scholar] [CrossRef]
- Nonyong, P.; Ekalaksananan, T.; Phanthanawiboon, S.; Aromseree, S.; Phadungsombat, J.; Nakayama, E.E.; Shioda, T.; Sawaswong, V.; Payungporn, S.; Thaewnongiew, K.; et al. Dengue virus in humans and mosquitoes and their molecular characteristics in northeastern Thailand 2016–2018. PLoS ONE 2021, 16, e0257460. [Google Scholar] [CrossRef] [PubMed]
- Phadungsombat, J.; Lin, M.Y.-C.; Srimark, N.; Yamanaka, A.; Nakayama, E.E.; Moolasart, V.; Suttha, P.; Shioda, T.; Uttayamakul, S. Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand. PLoS ONE 2018, 13, e0207220. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Phanthanawiboon, S.; Pambudi, S.; Omokoko, M.D.; Hanabara, K.; A-Nuegoonpipat, A.; Kamitani, W.; Ikuta, K.; Kurosu, T. Construction of a high-yield dengue virus by replacing nonstructural proteins 3–4B without increasing virulence. Biochem. Biophys. Res. Commun. 2018, 495, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Calisher, C.H.; Gubler, D.J.; Chang, G.J.; Vorndam, A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.-Y.; Chang, S.-F.; Kuo, Y.-C.; Yueh, Y.-Y.; Chien, L.-J.; Sue, C.-L.; Lin, T.-H.; Huang, J.-H. Development of Group- and Serotype-Specific One-Step SYBR Green I-Based Real-Time Reverse Transcription-PCR Assay for Dengue Virus. J. Clin. Microbiol. 2003, 41, 2408–2416. [Google Scholar] [CrossRef] [PubMed]
- Lequime, S.; Fontaine, A.; Gouilh, M.A.; Moltini-Conclois, I.; Lambrechts, L. Genetic Drift, Purifying Selection and Vector Genotype Shape Dengue Virus Intra-host Genetic Diversity in Mosquitoes. PLoS Genet. 2016, 12, e1006111. [Google Scholar] [CrossRef]
- Sullivan, D.G.; Bruden, D.; Deubner, H.; McArdle, S.; Chung, M.; Christensen, C.; Hennessy, T.; Homan, C.; Williams, J.; McMahon, B.J.; et al. Hepatitis C Virus Dynamics during Natural Infection Are Associated with Long-Term Histological Outcome of Chronic Hepatitis C Disease. J. Infect. Dis. 2007, 196, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Perelson, A.S.; Park, S.-C.; Leitner, T. Dynamic Correlation between Intrahost HIV-1 Quasispecies Evolution and Disease Progression. PLoS Comput. Biol. 2008, 4, e1000240. [Google Scholar] [CrossRef]
- Fitzsimmons, W.J.; Woods, R.J.; McCrone, J.T.; Woodman, A.; Arnold, J.J.; Yennawar, M.; Evans, R.; Cameron, C.E.; Lauring, A.S. A speed–fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol. 2018, 16, e2006459. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; López, C.B. Defective viral genomes are key drivers of the virus–host interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef]
- Ko, H.-Y.; Salem, G.M.; Chang, G.-J.J.; Chao, D.-Y. Application of Next-Generation Sequencing to Reveal How Evolutionary Dynamics of Viral Population Shape Dengue Epidemiology. Front. Microbiol. 2020, 11, 1371. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C. Patterns of Intra- and Interhost Nonsynonymous Variation Reveal Strong Purifying Selection in Dengue Virus. J. Virol. 2003, 77, 11296–11298. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; de Mendonça, M.L.; Rodrigues, C.D.D.S.; Fonseca, V.; Ribeiro, M.; Brandão, A.; da Cunha, R.V.; Dias, A.; Boas, L.S.V.; Felix, A.; et al. Dengue Virus Serotype 2 Intrahost Diversity in Patients with Different Clinical Outcomes. Viruses 2021, 13, 349. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zou, J.; Puttikhunt, C.; Yuan, Z.; Shi, P.-Y. Two Distinct Sets of NS2A Molecules Are Responsible for Dengue Virus RNA Synthesis and Virion Assembly. J. Virol. 2015, 89, 1298–1313. [Google Scholar] [CrossRef]
- Xie, X.; Gayen, S.; Kang, C.; Yuan, Z.; Shi, P.-Y. Membrane Topology and Function of Dengue Virus NS2A Protein. J. Virol. 2013, 87, 4609–4622. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, G.; García-Cordero, J.; León-Juárez, M.; Oza, G.; Tapia-Ramírez, J.; Villegas-Sepulveda, N.; Cedillo-Barrón, L. NS2A comprises a putative viroporin of Dengue virus 2. Virulence 2017, 8, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The flavivirus NS2B–NS3 protease–helicase as a target for antiviral drug development. Antivir. Res. 2015, 118, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.; Kuhn, R.J. Structural proteomics of dengue virus. Curr. Opin. Microbiol. 2008, 11, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Nasar, S.; Rashid, N.; Iftikhar, S. Dengue proteins with their role in pathogenesis, and strategies for developing an effective anti-dengue treatment: A review. J. Med. Virol. 2020, 92, 941–955. [Google Scholar] [CrossRef]
- Zhao, Y.; Soh, T.S.; Zheng, J.; Chan, K.W.K.; Phoo, W.W.; Lee, C.C.; Tay, M.; Swaminathan, K.; Cornvik, T.C.; Lim, S.P.; et al. A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication. PLoS Pathog. 2015, 11, e1004682. [Google Scholar] [CrossRef]
- Rothan, H.A.; Han, H.C.; Ramasamy, T.S.; Othman, S.; Rahman, N.A.; Yusof, R. Inhibition of dengue NS2B-NS3 protease and viral replication in Vero cells by recombinant retrocyclin-1. BMC Infect. Dis. 2012, 12, 314. [Google Scholar] [CrossRef]
- Chambers, T.J.; Nestorowicz, A.; Rice, C.M. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: Determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J. Virol. 1995, 69, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Green, A.M.; Beatty, P.R.; Hadjilaou, A.; Harris, E. Innate Immunity to Dengue Virus Infection and Subversion of Antiviral Responses. J. Mol. Biol. 2014, 426, 1148–1160. [Google Scholar] [CrossRef]
- Mlera, L.; Melik, W.; Bloom, M.E. The role of viral persistence in flavivirus biology. Pathog. Dis. 2014, 71, 137–163. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vargas, I.; Scott, J.C.; Poole-Smith, B.K.; Franz, A.W.E.; Barbosa-Solomieu, V.; Wilusz, J.; Olson, K.E.; Blair, C.D. Dengue Virus Type 2 Infections of Aedes aegypti Are Modulated by the Mosquito’s RNA Interference Pathway. PLoS Pathog. 2009, 5, e1000299. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Jordán, J.L.; Sánchez-Burgos, G.G.; Laurent-Rolle, M.; García-Sastre, A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 2003, 100, 14333–14338. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Chen, H.B.; Wang, X.J.; Huang, H.; Khromykh, A.A. Analysis of Adaptive Mutations in Kunjin Virus Replicon RNA Reveals a Novel Role for the Flavivirus Nonstructural Protein NS2A in Inhibition of Beta Interferon Promoter-Driven Transcription. J. Virol. 2004, 78, 12225–12235. [Google Scholar] [CrossRef]
- Baril, M.; Racine, M.E.; Penin, F.; Lamarre, D. MAVS Dimer Is a Crucial Signaling Component of Innate Immunity and the Target of Hepatitis C Virus NS3/4A Protease. J. Virol. 2009, 83, 1299–1311. [Google Scholar] [CrossRef]
- Hudopisk, N.; Korva, M.; Janet, E.; Simetinger, M.; Grgič-Vitek, M.; Gubensek, J.; Natek, V.; Kraigher, A.; Strle, F.; Avšič-Županc, T. Tick-borne Encephalitis Associated with Consumption of Raw Goat Milk, Slovenia, 2012. Emerg. Infect. Dis. 2013, 19, 806–808. [Google Scholar] [CrossRef]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus Entry Receptors: An Update. Viruses 2013, 6, 69–88. [Google Scholar] [CrossRef]

| Name | nt c | aa d | Primer Sequence (5′→3′) |
|---|---|---|---|
| ∆NS2A-F a | A3721G | K81R | CCATCATGGCAGTGTTCAGGATGTCAC |
| ∆NS2A-R b | CGTATCCTGGTGACATCCTGAACACTG | ||
| ∆NS3-F a | A4843G | K107R | GAACCAGGAAAAAATCCGAGACATGTCC |
| ∆NS3-R b | GTTTCGTTTGGACATGTCTCGGA | ||
| ∆NS5-F a | A9249G | I563V | GAT GGCCCCTCATCATAAAGTCCTAGC |
| ∆NS5-R b | GAAAATGGCTTTAGCTAGGACTTTATGATG |
| Nucleotide a | Amino Acid b | Genome Region/Gene | Substitution Frequency (%) | |
|---|---|---|---|---|
| Mosquito (n = 2) | Human (n = 11) | |||
| G186A | V29M | C | 100 | 45.5 |
| A657G | T73A | prM | 100 | 90.9 |
| G1602A | A222T | E | 100 | 45.5 |
| C1637T | Y233H | E | 100 | 45.5 |
| T2223C | F429L | E | 100 | 90.9 |
| T2437C | V5A | NS1 | 100 | 81.8 |
| C2717A | T98S | NS1 | 100 | 90.9 |
| T2746C | V108A | NS1 | 50 | 0 |
| G2805A | A128T | NS1 | 100 | 90.9 |
| C2880T | L153F | NS1 | 100 | 90.9 |
| T3159G | S246A | NS1 | 100 | 81.8 |
| A3721G * | K81R | NS2A | 100 | 18.2 |
| T3980G | I167M | NS2A | 50 | 0 |
| C4555T | A11V | NS3 | 50 | 0 |
| A4574G | A17T | NS3 | 50 | 0 |
| A4643G | I40V | NS3 | 50 | 0 |
| A4843G * | K107R | NS3 | 100 | 0 |
| G5484A | A321T | NS3 | 100 | 81.8 |
| A5828T | P435T | NS3 | 100 | 81.8 |
| A5962G | K480R | NS3 | 100 | 81.8 |
| C8368 | T269I | NS5 | 100 | 90.9 |
| A9249G * | I563V | NS5 | 100 | 45.5 |
| T9387A | L609M | NS5 | 100 | 63.6 |
| G9467T | Q635H | NS5 | 100 | 90.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nonyong, P.; Ekalaksananan, T.; Phanthanawiboon, S.; Overgaard, H.J.; Alexander, N.; Thaewnongiew, K.; Sawaswong, V.; Nimsamer, P.; Payungporn, S.; Phadungsombat, J.; et al. Intrahost Genetic Diversity of Dengue Virus in Human Hosts and Mosquito Vectors under Natural Conditions Which Impact Replicative Fitness In Vitro. Viruses 2023, 15, 982. https://doi.org/10.3390/v15040982
Nonyong P, Ekalaksananan T, Phanthanawiboon S, Overgaard HJ, Alexander N, Thaewnongiew K, Sawaswong V, Nimsamer P, Payungporn S, Phadungsombat J, et al. Intrahost Genetic Diversity of Dengue Virus in Human Hosts and Mosquito Vectors under Natural Conditions Which Impact Replicative Fitness In Vitro. Viruses. 2023; 15(4):982. https://doi.org/10.3390/v15040982
Chicago/Turabian StyleNonyong, Patcharaporn, Tipaya Ekalaksananan, Supranee Phanthanawiboon, Hans J. Overgaard, Neal Alexander, Kesorn Thaewnongiew, Vorthon Sawaswong, Pattaraporn Nimsamer, Sunchai Payungporn, Juthamas Phadungsombat, and et al. 2023. "Intrahost Genetic Diversity of Dengue Virus in Human Hosts and Mosquito Vectors under Natural Conditions Which Impact Replicative Fitness In Vitro" Viruses 15, no. 4: 982. https://doi.org/10.3390/v15040982
APA StyleNonyong, P., Ekalaksananan, T., Phanthanawiboon, S., Overgaard, H. J., Alexander, N., Thaewnongiew, K., Sawaswong, V., Nimsamer, P., Payungporn, S., Phadungsombat, J., Nakayama, E. E., Shioda, T., & Pientong, C. (2023). Intrahost Genetic Diversity of Dengue Virus in Human Hosts and Mosquito Vectors under Natural Conditions Which Impact Replicative Fitness In Vitro. Viruses, 15(4), 982. https://doi.org/10.3390/v15040982







