In Vitro Effect on Piglet Gut Microbiota and In Vivo Assessment of Newly Isolated Bacteriophages against F18 Enterotoxigenic Escherichia coli (ETEC)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Phage Isolation
2.3. Phage Titration and Host Range
2.4. Temperature and pH Stability
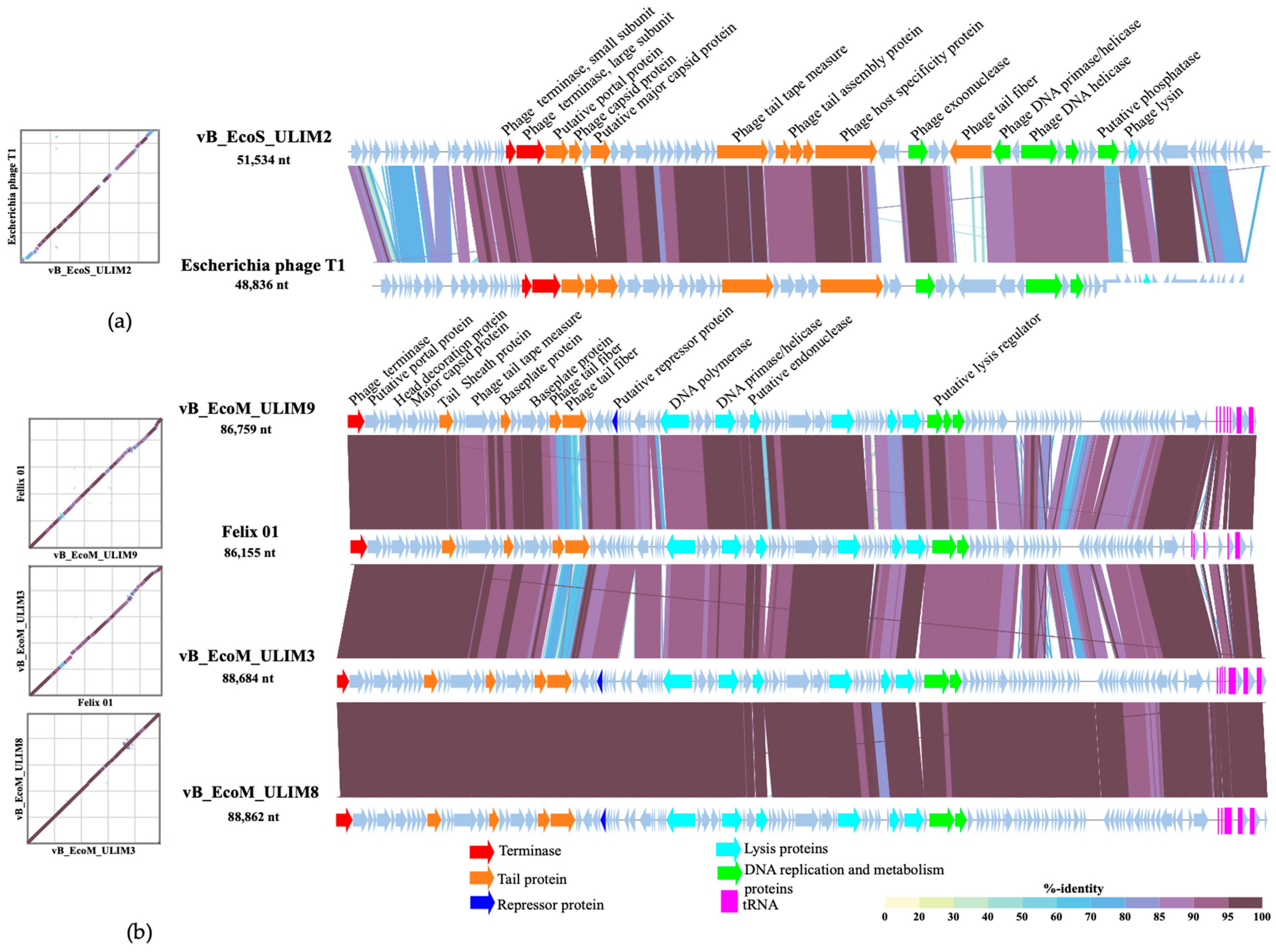
2.5. Genomic Analysis
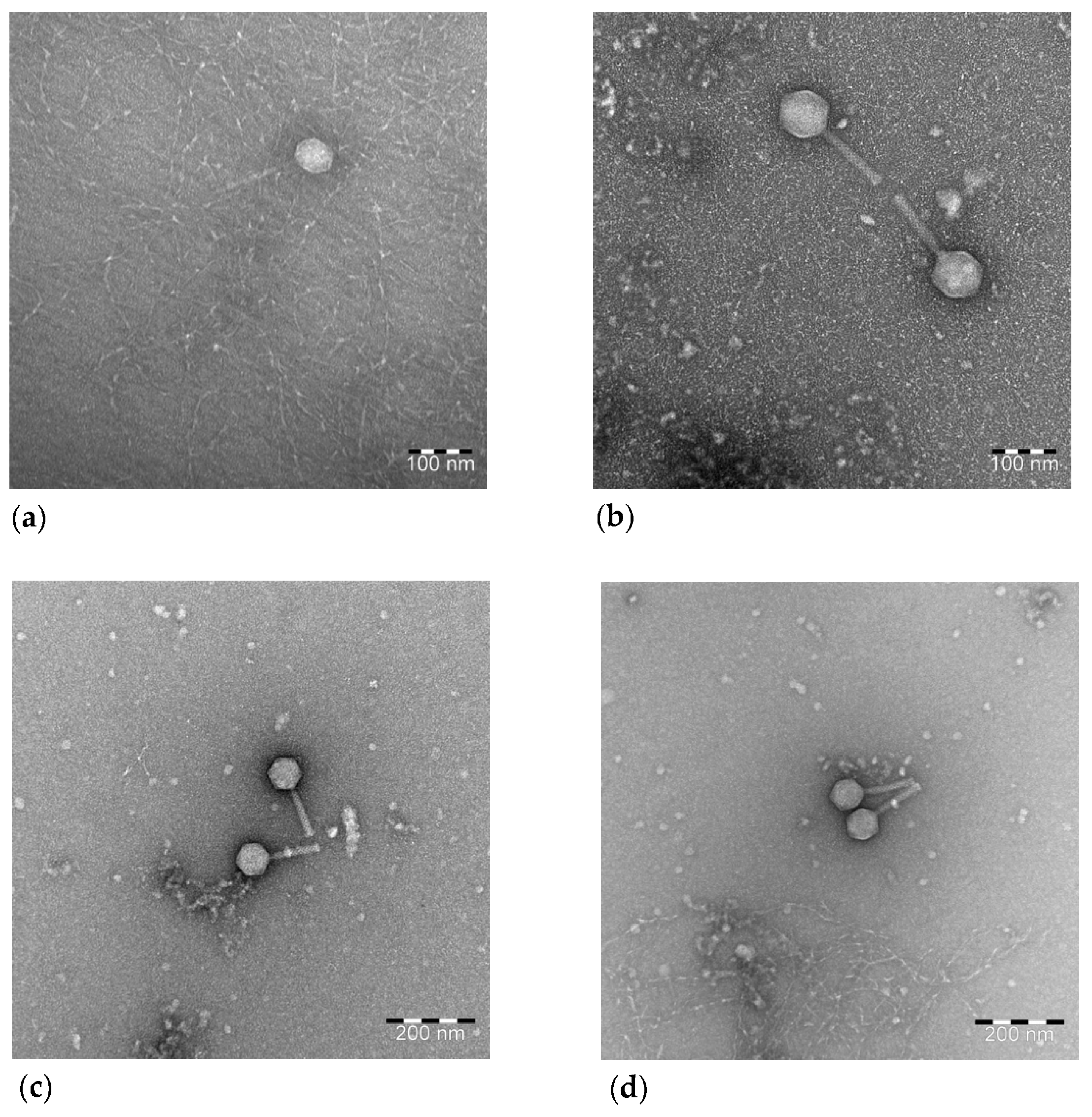
2.6. Transmission Electron Microscopy
2.7. Galleria Mellonella Larvae Model
2.8. Intestinal Batches Model
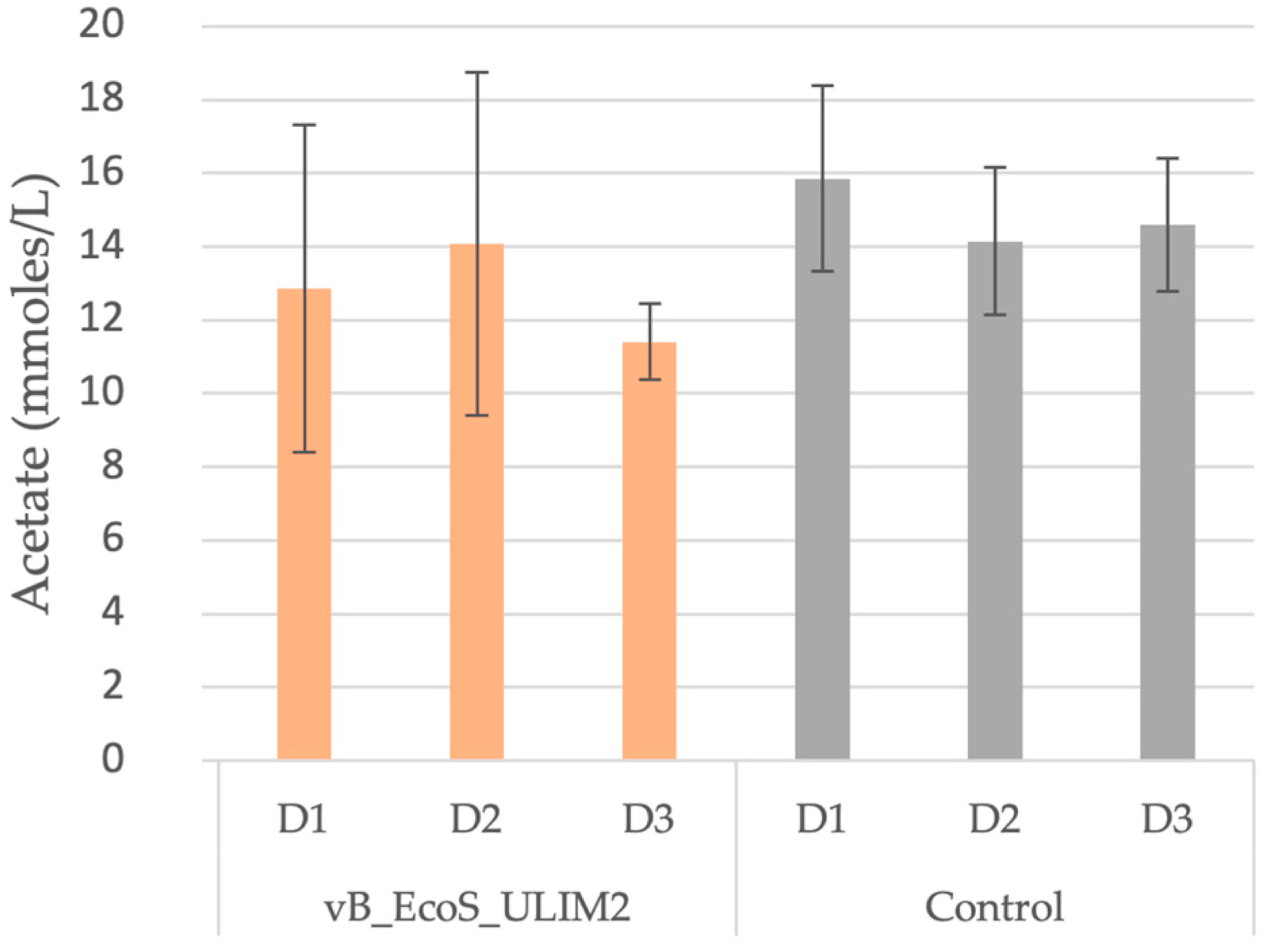
2.9. Short-Chain Fatty Acids Analysis
2.10. Metagenetic Analysis
3. Results
3.1. Phage Isolation
3.2. Host Range
3.3. Temperature and pH Stabilities
3.4. Genomic Analysis
3.5. Transmission Electron Microscopy
3.6. Galleria Mellonella Larvae Experiments
3.7. Short-Chain Fatty Acid and Metagenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buuck, S.; Smith, K.; Fowler, R.C.; Cebelinski, E.; Lappi, V.; Boxrud, D.; Medus, C. Epidemiology of Enterotoxigenic Escherichia Coli Infection in Minnesota, 2016–2017. Epidemiol. Infect. 2020, 148, e206. [Google Scholar] [CrossRef]
- Croxen, M.A.; Finlay, B.B. Molecular Mechanisms of Escherichia Coli Pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef]
- Pakbin, B.; Brück, W.M.; Rossen, J.W.A. Virulence Factors of Enteric Pathogenic Escherichia Coli: A Review. Int. J. Mol. Sci. 2021, 22, 9922. [Google Scholar] [CrossRef] [PubMed]
- Luise, D.; Lauridsen, C.; Bosi, P.; Trevisi, P. Methodology and Application of Escherichia Coli F4 and F18 Encoding Infection Models in Post-Weaning Pigs. J. Anim. Sci. Biotechnol. 2019, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.J.; Karriker, L.A.; Ramirez, A.; Schwartz, K.J.; Stevenson, G.W.; Zhang, J. Colibacillosis, 11th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; ISBN 978-1-119-35085-9. [Google Scholar]
- Bin, P.; Tang, Z.; Liu, S.; Chen, S.; Xia, Y.; Liu, J.; Wu, H.; Zhu, G. Intestinal Microbiota Mediates Enterotoxigenic Escherichia Coli-Induced Diarrhea in Piglets. BMC Vet. Res. 2018, 14, 385. [Google Scholar] [CrossRef]
- Luppi, A.; Gibellini, M.; Gin, T.; Vangroenweghe, F.; Vandenbroucke, V.; Bauerfeind, R.; Bonilauri, P.; Labarque, G.; Hidalgo, Á. Prevalence of Virulence Factors in Enterotoxigenic Escherichia Coli Isolated from Pigs with Post-Weaning Diarrhoea in Europe. Porc. Health Manag. 2016, 2, 20. [Google Scholar] [CrossRef]
- Sinha, R.; Sahoo, N.R.; Shrivastava, K.; Kumar, P.; Qureshi, S.; De, U.K.; Kumar, A.; Kumar, G.V.P.P.S.R.; Bhushan, B. Resistance to ETEC F4/F18–Mediated Piglet Diarrhoea: Opening the Gene Black Box. Trop. Anim. Health Prod. 2019, 51, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post Weaning Diarrhea in Pigs: Risk Factors and Non-Colistin-Based Control Strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Bergeron, N.; Beauchamp, G.; Laurent-Lewandowski, S.; Fairbrother, J.M.; Letellier, A. In Vivo Therapeutic Efficacy and Pharmacokinetics of Colistin Sulfate in an Experimental Model of Enterotoxigenic Escherichia Coli Infection in Weaned Pigs. Vet. Res. 2016, 47, 58. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Laird, T.J.; Abraham, S.; Jordan, D.; Pluske, J.R.; Hampson, D.J.; Trott, D.J.; O’Dea, M. Porcine Enterotoxigenic Escherichia Coli: Antimicrobial Resistance and Development of Microbial-Based Alternative Control Strategies. Vet. Microbiol. 2021, 258, 109117. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Gu, M.J.; Kye, Y.C.; Ju, Y.J.; Hong, R.; Ju, D.B.; Pyung, Y.J.; Han, S.H.; Park, B.C.; Yun, C.H. Bacteriophage EK99P-1 Alleviates Enterotoxigenic Escherichia Coli K99-Induced Barrier Dysfunction and Inflammation. Sci. Rep. 2022, 12, 941. [Google Scholar] [CrossRef]
- Cha, S.B.; Yoo, A.N.; Lee, W.J.; Shin, M.K.; Jung, M.H.; Shin, S.W.; Cho, Y.W.; Yoo, H.S. Effect of Bacteriophage in Enterotoxigenic Escherichia Coli (ETEC) Infected Pigs. J. Vet. Med. Sci. 2012, 74, 1037–1039. [Google Scholar] [CrossRef]
- Lee, C.Y.; Kim, S.J.; Park, B.C.; Han, J.H. Effects of Dietary Supplementation of Bacteriophages against Enterotoxigenic Escherichia Coli (ETEC) K88 on Clinical Symptoms of Post-Weaning Pigs Challenged with the ETEC Pathogen. J. Anim. Physiol. Anim. Nutr. 2017, 101, 88–95. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, Stabilisation and Encapsulation of Bacteriophage for Phage Therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef] [PubMed]
- Barnoy, S.; Gancz, H.; Zhu, Y.; Honnold, C.L.; Zurawski, D.V.; Venkatesan, M.M. The Galleria Mellonella Larvae as an In Vivo Model for Evaluation of Shigella Virulence. Gut Microbes 2017, 8, 335–350. [Google Scholar] [CrossRef]
- Cutuli, M.A.; Petronio Petronio, G.; Vergalito, F.; Magnifico, I.; Pietrangelo, L.; Venditti, N.; Di Marco, R. Galleria Mellonella as a Consolidated In Vivo Model Hosts: New Developments in Antibacterial Strategies and Novel Drug Testing. Virulence 2019, 10, 527–541. [Google Scholar] [CrossRef]
- Antoine, C.; Laforêt, F.; Blasdel, B.; Fall, A.; Duprez, J.N.; Mainil, J.; Delcenserie, V.; Thiry, D. In Vitro Characterization and In Vivo Efficacy Assessment in Galleria Mellonella Larvae of Newly Isolated Bacteriophages against Escherichia Coli K1. Viruses 2021, 13, 2005. [Google Scholar] [CrossRef]
- Petronio Petronio, G.; Cutuli, M.A.; Magnifico, I.; Venditti, N.; Pietrangelo, L.; Vergalito, F.; Pane, A.; Scapagnini, G.; Di Marco, R. In Vitro and In Vivo Biological Activity of Berberine Chloride against Uropathogenic E. coli Strains Using Galleria Mellonella as a Host Model. Molecules 2020, 25, 5010. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Ramesh, N. Isolation, Characterization and In Vivo Efficacy of Escherichia Phage MyPSH1131. PLoS ONE 2018, 13, e0206278. [Google Scholar] [CrossRef]
- Goya-Jorge, E.; Gonza, I.; Bondue, P.; Douny, C.; Taminiau, B.; Daube, G.; Scippo, M.-L.; Delcenserie, V. Human Adult Microbiota in a Static Colon Model: AhR Transcriptional Activity at the Crossroads of Host–Microbe Interaction. Foods 2022, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Neckermann, K.; Claus, G.; De Baere, S.; Antonissen, G.; Lebrun, S.; Gemmi, C.; Taminiau, B.; Douny, C.; Scippo, M.L.; Schatzmayr, D.; et al. The Efficacy and Effect on Gut Microbiota of an Aflatoxin Binder and a Fumonisin Esterase Using an In Vitro Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). Food Res. Int. 2021, 145, 110395. [Google Scholar] [CrossRef]
- Dufourny, S.; Everaert, N.; Lebrun, S.; Douny, C.; Scippo, M.L.; Li, B.; Taminiau, B.; Marzorati, M.; Wavreille, J.; Froidmont, E.; et al. Baby-SPIME: A Dynamic In Vitro Piglet Model Mimicking Gut Microbiota during the Weaning Process. J. Microbiol. Methods 2019, 167, 105735. [Google Scholar] [CrossRef]
- Tóth, I.; Oswald, E.; Mainil, J.G.; Awad-Masalmeh, M.; Nagy, B. Characterization of Intestinal Cnf1+ Escherichia Coli from Weaned Pigs. Int. J. Med. Microbiol. 2000, 290, 539–542. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Tolstoy, I.; Kropinski, A.M. Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation. PHAGE 2021, 2, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Meier, A.; Söding, J. Automatic Prediction of Protein 3D Structures by Probabilistic Multi-Template Homology Modeling. PLoS Comput. Biol. 2015, 11, e1004343. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a Mucosal Environment in a Dynamic Gut Model Results in a More Representative Colonization by Lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Grootaert, C.; Marzorati, M.; Possemiers, S.; Verstraete, W.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.; et al. Microbial Community Development in a Dynamic Gut Model Is Reproducible, Colon Region Specific, and Selective for Bacteroidetes and Clostridium Cluster IX. Appl. Environ. Microbiol. 2010, 76, 5237–5246. [Google Scholar] [CrossRef] [PubMed]
- Bondue, P.; Lebrun, S.; Taminiau, B.; Everaert, N.; LaPointe, G.; Crevecoeur, S.; Daube, G.; Delcenserie, V. A Toddler SHIME® Model to Study Microbiota of Young Children. FEMS Microbiol. Lett. 2020, 367, fnaa135. [Google Scholar] [CrossRef]
- Bondue, P.; Lebrun, S.; Taminiau, B.; Everaert, N.; LaPointe, G.; Hendrick, C.; Gaillez, J.; Crèvecoeur, S.; Daube, G.; Delcenserie, V. Effect of Bifidobacterium Crudilactis and 3′-Sialyllactose on the Toddler Microbiota Using the SHIME® Model. Food Res. Int. 2020, 138, 109755. [Google Scholar] [CrossRef] [PubMed]
- Barron-Montenegro, R.; García, R.; Dueñas, F.; Rivera, D.; Opazo-Capurro, A.; Erickson, S.; Moreno-Switt, A.I. Comparative Analysis of Felixounavirus Genomes Including Two New Members of the Genus That Infect Salmonella Infantis. Antibiotics 2021, 10, 806. [Google Scholar] [CrossRef]
- Hua, Y.; Luo, T.; Yang, Y.; Dong, D.; Wang, R.; Wang, Y.; Xu, M.; Guo, X.; Hu, F.; He, P. Phage Therapy as a Promising New Treatment for Lung Infection Caused by Carbapenem-Resistant Acinetobacter Baumannii in Mice. Front. Microbiol. 2018, 8, 2659. [Google Scholar] [CrossRef] [PubMed]
- Jennes, S.; Merabishvili, M.; Soentjens, P.; Pang, K.W.; Rose, T.; Keersebilck, E.; Soete, O.; François, P.M.; Teodorescu, S.; Verween, G.; et al. Use of Bacteriophages in the Treatment of Colistin-Only-Sensitive Pseudomonas Aeruginosa Septicaemia in a Patient with Acute Kidney Injury—A Case Report. Crit. Care 2017, 21, 2016–2018. [Google Scholar] [CrossRef]
- Regeimbal, J.M.; Jacobs, A.C.; Corey, B.W.; Henry, M.S.; Thompson, M.G.; Pavlicek, R.L.; Quinones, J.; Hannah, R.M.; Ghebremedhin, M.; Crane, N.J.; et al. Personalized Therapeutic Cocktail of Wild Environmental Phages Rescues Mice from Acinetobacter Baumannii Wound Infections. Antimicrob. Agents Chemother. 2016, 60, 5806–5816. [Google Scholar] [CrossRef]
- Ngassam-Tchamba, C.; Duprez, J.N.; Fergestad, M.; De Visscher, A.; L’Abee-Lund, T.; De Vliegher, S.; Wasteson, Y.; Touzain, F.; Blanchard, Y.; Lavigne, R.; et al. In Vitro and In Vivo Assessment of Phage Therapy against Staphylococcus Aureus Causing Bovine Mastitis. J. Glob. Antimicrob. Resist. 2020, 22, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.G.; Jordan, D.; Chapman, T.A.; Chin, J.J.C.; Barton, M.D.; Do, T.N.; Fahy, V.A.; Fairbrother, J.M.; Trott, D.J. Antimicrobial Resistance and Virulence Gene Profiles in Multi-Drug Resistant Enterotoxigenic Escherichia Coli Isolated from Pigs with Post-Weaning Diarrhoea. Vet. Microbiol. 2010, 145, 299–307. [Google Scholar] [CrossRef]
- Vu-Khac, H.; Holoda, E.; Pilipčinec, E. Distribution of Virulence Genes in Escherichia Coli Strains Isolated from Diarrhoeic Piglets in the Slovak Republic. J. Vet. Med. Ser. B Infect. Dis. Vet. Public Health 2004, 51, 343–347. [Google Scholar] [CrossRef]
- Wei, C.; Zhao, X. Induction of Viable but Nonculturable Escherichia Coli O157:H7 by Low Temperature and Its Resuscitation. Front. Microbiol. 2018, 9, 2728. [Google Scholar] [CrossRef]
- Górski, A.; Miedzybrodzki, R.; Borysowski, J. Phage Therapy: A Practical Approach; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Assafiri, O.; Song, A.A.L.; Tan, G.H.; Hanish, I.; Hashim, A.M.; Yusoff, K. Klebsiella Virus UPM2146 Lyses Multiple Drug-Resistant Klebsiella Pneumoniae In Vitro and In Vivo. PLoS ONE 2021, 16, e0245354. [Google Scholar] [CrossRef]
- Merabishvili, M.; Vandenheuvel, D.; Kropinski, A.M.; Mast, J.; De Vos, D.; Verbeken, G.; Noben, J.P.; Lavigne, R.; Vaneechoutte, M.; Pirnay, J.P. Characterization of Newly Isolated Lytic Bacteriophages Active against Acinetobacter Baumannii. PLoS ONE 2014, 9, e104853. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the Intriguing Presence of TRNAs in Phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.H.; Michie, K.A.; Goh, Y.F.; Noorian, P.; Kjelleberg, S.; Duggin, I.G.; McDougald, D.; Rice, S.A. The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas Aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression. Viruses 2021, 13, 1614. [Google Scholar] [CrossRef]
- Jeon, J.; Ryu, C.M.; Lee, J.Y.; Park, J.H.; Yong, D.; Lee, K. In Vivo Application of Bacteriophage as a Potential Therapeutic Agent to Control OXA-66-like Carbapenemase-Producing Acinetobacter Baumannii Strains Belonging to Sequence Type 357. Appl. Environ. Microbiol. 2016, 82, 4200–4208. [Google Scholar] [CrossRef]
- Cao, F.; Wang, X.; Wang, L.; Li, Z.; Che, J.; Wang, L.; Li, X.; Cao, Z.; Zhang, J.; Jin, L.; et al. Evaluation of the Efficacy of a Bacteriophage in the Treatment of Pneumonia Induced by Multidrug Resistance Klebsiella Pneumoniae in Mice. BioMed. Res. Int. 2015, 2015, 752930. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F. Escherichia Coli O157:H7 Bacteriophage F241 Isolated from an Industrial Cucumber Fermentation at High Acidity and Salinity. Front. Microbiol. 2015, 6, 67. [Google Scholar] [CrossRef]
- Williams, C.F.; Walton, G.E.; Jiang, L.; Plummer, S.; Garaiova, I.; Gibson, G.R. Comparative Analysis of Intestinal Tract Models. Annu. Rev. Food Sci. Technol. 2015, 6, 329–350. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the Effects of Diet on Bacterial Metabolism in the Large Intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 121, pp. 91–119. ISBN 978-0-12-800100-4. [Google Scholar]
- Bernalier-Donadille, A. Fermentative Metabolism by the Human Gut Microbiota. Gastroentérologie Clin. Biol. 2010, 34, S16–S22. [Google Scholar] [CrossRef]
- Febvre, H.; Rao, S.; Gindin, M.; Goodwin, N.; Finer, E.; Vivanco, J.; Lu, S.; Manter, D.; Wallace, T.; Weir, T. PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K. The Intestinal Microbiota and Its Role in Human Health and Disease. J. Med. Invest. 2016, 63, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Gresse, R.; Chaucheyras-Durand, F.; Fleury, M.A.; Van de Wiele, T.; Forano, E.; Blanquet-Diot, S. Gut Microbiota Dysbiosis in Postweaning Piglets: Understanding the Keys to Health. Trends Microbiol. 2017, 25, 851–873. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-Derived Nitrate Boosts Growth of E. coli in the Inflamed Gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Dufourny, S.; Everaert, N.; Lebrun, S.; Didelez, M.; Wavreille, J.; Froidmont, E.; Rondia, P.; Delcenserie, V. Oxygen as a Key Parameter in In Vitro Dynamic and Multi-Compartment Models to Improve Microbiome Studies of the Small Intestine? Food Res. Int. 2020, 133, 109127. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, R.R.; Trinh, J.T.; Bomholtz, L.; Brok-Lauridsen, S.K.; Sulakvelidze, A.; Nielsen, D.S. A Bacteriophage Cocktail Significantly Reduces Listeria Monocytogenes without Deleterious Impact on the Commensal Gut Microbiota under Simulated Gastrointestinal Conditions. Viruses 2022, 14, 190. [Google Scholar] [CrossRef]
- Grubb, D.S.; Wrigley, S.D.; Freedman, K.E.; Wei, Y.; Vazquez, A.R.; Trotter, R.E.; Wallace, T.C.; Johnson, S.A.; Weir, T.L. PHAGE-2 Study: Supplemental Bacteriophages Extend Bifidobacterium Animalis Subsp. Lactis BL04 Benefits on Gut Health and Microbiota in Healthy Adults. Nutrients 2020, 12, 2474. [Google Scholar] [CrossRef]
- Dissanayake, U.; Ukhanova, M.; Moye, Z.D.; Sulakvelidze, A.; Mai, V. Bacteriophages Reduce Pathogenic Escherichia Coli Counts in Mice Without Distorting Gut Microbiota. Front. Microbiol. 2019, 10, 1984. [Google Scholar] [CrossRef]
- Pinto, G.; Shetty, S.A.; Zoetendal, E.G.; Gonçalves, R.F.S.; Pinheiro, A.C.; Almeida, C.; Azeredo, J.; Smidt, H. An In Vitro Fermentation Model to Study the Impact of Bacteriophages Targeting Shiga Toxin-Encoding Escherichia Coli on the Colonic Microbiota. NPJ Biofilms Microbiomes 2022, 8, 74. [Google Scholar] [CrossRef]
- Tetz, G.V.; Ruggles, K.V.; Zhou, H.; Heguy, A.; Tsirigos, A.; Tetz, V. Bacteriophages as Potential New Mammalian Pathogens. Sci. Rep. 2017, 7, 7043. [Google Scholar] [CrossRef]
- Tetz, G.; Tetz, V. Bacteriophage Infections of Microbiota Can Lead to Leaky Gut in an Experimental Rodent Model. Gut Pathog. 2016, 8, 33. [Google Scholar] [CrossRef]
- Laforêt, F.; Antoine, C.; Lebrun, S.; Gonza, I.; Goya-Jorge, E.; Douny, C.; Duprez, J.-N.; Scippo, M.-L.; Taminiau, B.; Daube, G.; et al. Impact Assessment of VB_KpnP_K1-ULIP33 Bacteriophage on the Human Gut Microbiota Using a Dynamic In Vitro Model. Viruses 2023, 15, 719. [Google Scholar] [CrossRef] [PubMed]
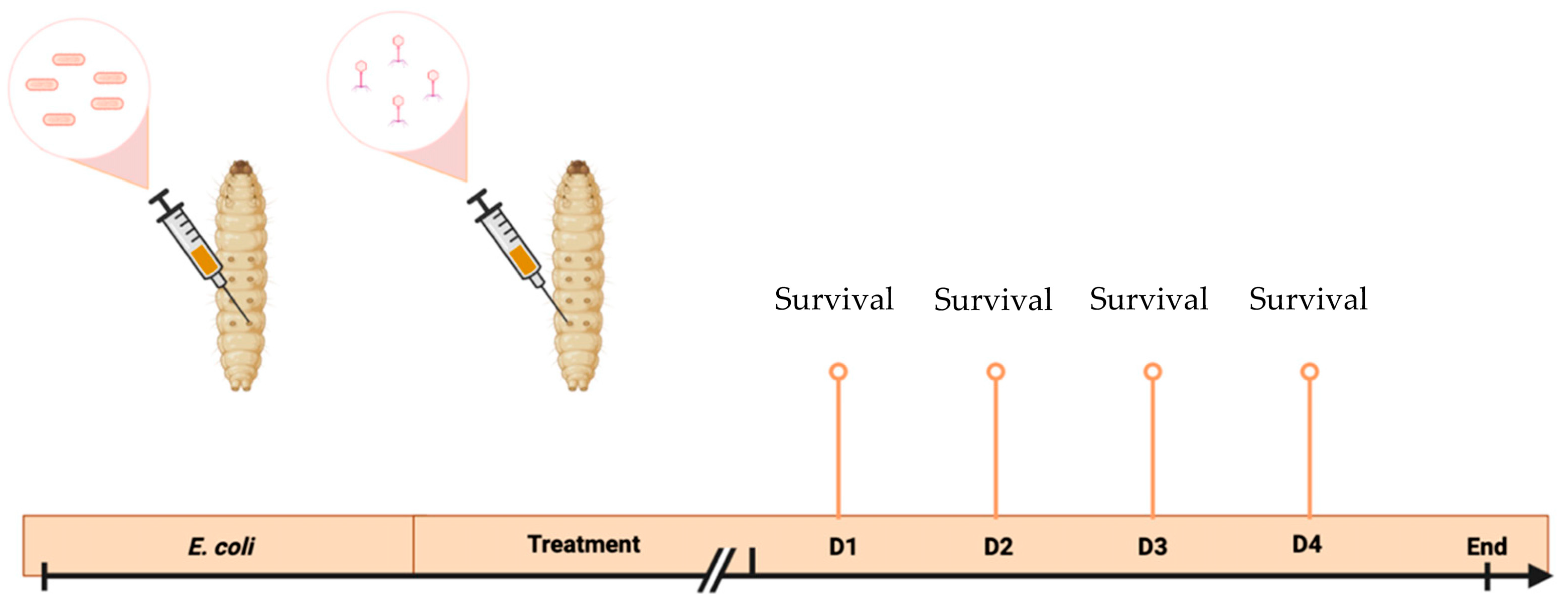




| Groups | 1st Injection (10 μL) | 2nd Injection (10 μL) | |
|---|---|---|---|
| 1 | PBS + PBS | PBS | PBS |
| 2 | A-I-210 + PBS | A-I-210: 104 CFU | PBS |
| 3 | PBS + phage 1 MOI 100 | PBS | Phage 1: 106 PFU |
| 4 | A-I-210 + phage 1 MOI 10 | A-I-210: 104 CFU | Phage 1: 105 PFU |
| 5 | A-I-210 + phage 1 MOI 100 | A-I-210: 104 CFU | Phage 1: 106 PFU |
| Bacteriophages | |||||
|---|---|---|---|---|---|
| Fimbriae type | E. coli strains | vB_EcoS-ULIM2 | vB_EcoM-ULIM3 | vB_EcoM-ULIM8 | vB_EcoM-ULIM9 |
| F4 | E65 | N | 3 | 3 | 3 |
| G7 | N | N | N | N | |
| G205 | N | 1 | 1 | N | |
| G491 | N | N | 3 | N | |
| Abbotstown | N | N | N | N | |
| 24KP88 | N | N | N | N | |
| 107KP88 | N | N | N | N | |
| F18 | A-I-9 | N | 1 | 1 | 1 |
| A-I-85 | N | 1 | 1 | 1 | |
| A-I-136 | N | N | N | N | |
| A-I-137 | N | 1 | 1 | N | |
| A-I-154 | 1 | 1 | 1 | 3 | |
| A-I-164 | 1 | N | N | N | |
| A-I-210 | 1 | 1 | 1 | 1 | |
| A-I-219 | 1 | 1 | 1 | 1 | |
| A-I-220 | 2 | 1 | 1 | 1 | |
| A-I-222 | 1 | 1 | 1 | 1 | |
| A-II-37 | N | 1 | 3 | N | |
| A-II-40 | N | 1 | 1 | 1 | |
| Chao1 | Simpson | Shannon | |
|---|---|---|---|
| Control | 169 ± 81.63 | 0.49 ± 0.27 | 1.40 ± 0.64 |
| vB_EcoS_ULIM2 | 108 ± 33.03 | 0.25 ± 0.30 | 2.54 ± 1.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navez, M.; Antoine, C.; Laforêt, F.; Goya-Jorge, E.; Douny, C.; Scippo, M.-L.; Vermeersch, M.; Duprez, J.-N.; Daube, G.; Mainil, J.; et al. In Vitro Effect on Piglet Gut Microbiota and In Vivo Assessment of Newly Isolated Bacteriophages against F18 Enterotoxigenic Escherichia coli (ETEC). Viruses 2023, 15, 1053. https://doi.org/10.3390/v15051053
Navez M, Antoine C, Laforêt F, Goya-Jorge E, Douny C, Scippo M-L, Vermeersch M, Duprez J-N, Daube G, Mainil J, et al. In Vitro Effect on Piglet Gut Microbiota and In Vivo Assessment of Newly Isolated Bacteriophages against F18 Enterotoxigenic Escherichia coli (ETEC). Viruses. 2023; 15(5):1053. https://doi.org/10.3390/v15051053
Chicago/Turabian StyleNavez, Margaux, Céline Antoine, Fanny Laforêt, Elizabeth Goya-Jorge, Caroline Douny, Marie-Louise Scippo, Marjorie Vermeersch, Jean-Noël Duprez, Georges Daube, Jacques Mainil, and et al. 2023. "In Vitro Effect on Piglet Gut Microbiota and In Vivo Assessment of Newly Isolated Bacteriophages against F18 Enterotoxigenic Escherichia coli (ETEC)" Viruses 15, no. 5: 1053. https://doi.org/10.3390/v15051053






