Analyses of Early ZIKV Genomes Are Consistent with Viral Spread from Northeast Brazil to the Americas
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Nucleic Acid Isolation and RT-qPCR
2.3. Complete Genome MinION Nanopore Sequencing
2.4. Generation of Consensus Sequences from Nanopore
2.5. Collation of Sequence Dataset
2.6. Maximum Likelihood Analysis and Temporal Signal Estimation
2.7. Molecular Clock Phylogenetic Analysis
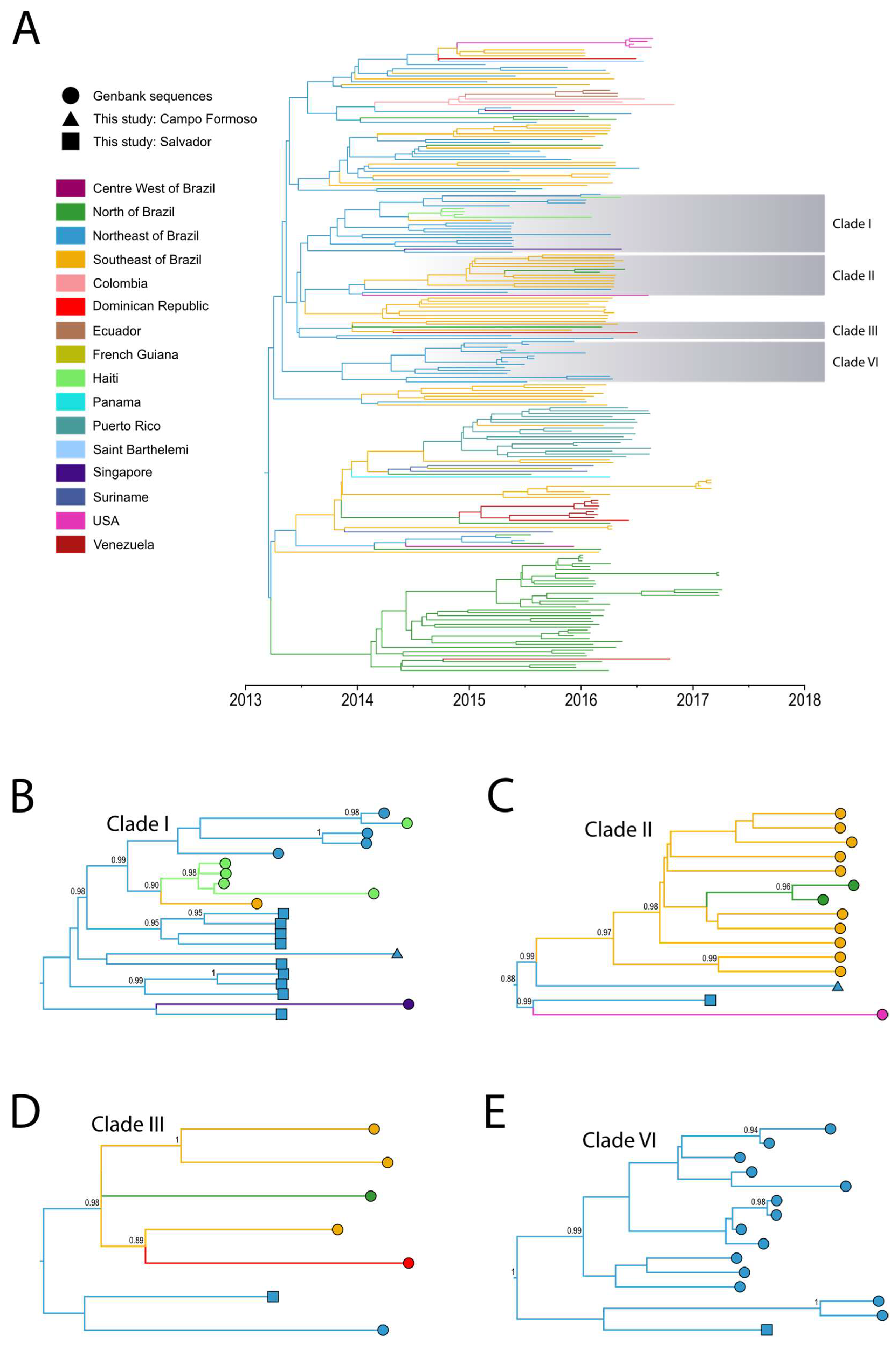
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Broutet, N.; Krauer, F.; Riesen, M.; Khalakdina, A.; Almiron, M.; Aldighieri, S.; Espinal, M.; Low, N.; Dye, C. Zika Virus as a Cause of Neurologic Disorders. N. Engl. J. Med. 2016, 374, 1506–1509. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.-J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the Genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.-J.J. Full-Length Sequencing and Genomic Characterization of Bagaza, Kedougou, and Zika Viruses. Arch. Virol. 2007, 152, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Simón, D.; Fajardo, A.; Moreno, P.; Moratorio, G.; Cristina, J. An Evolutionary Insight into Zika Virus Strains Isolated in the Latin American Region. Viruses 2018, 10, 698. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and Serologic Properties of Zika Virus Associated with an Epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef]
- Aubry, M.; Teissier, A.; Huart, M.; Merceron, S.; Vanhomwegen, J.; Roche, C.; Vial, A.-L.; Teururai, S.; Sicard, S.; Paulous, S.; et al. Zika Virus Seroprevalence, French Polynesia, 2014–2015. Emerg. Infect. Dis. 2017, 23, 669–672. [Google Scholar] [CrossRef]
- Zanluca, C.; de Melo, V.C.A.; Mosimann, A.L.P.; dos Santos, G.I.V.; dos Santos, C.N.D.; Luz, K. First Report of Autochthonous Transmission of Zika Virus in Brazil. Mem. Inst. Oswaldo Cruz. 2015, 110, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg. Infect. Dis. 2015, 21, 1885–1886. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Ladner, J.T.; Kraemer, M.U.G.; Dudas, G.; Tan, A.L.; Gangavarapu, K.; Wiley, M.R.; White, S.; Thézé, J.; Magnani, D.M.; et al. Genomic Epidemiology Reveals Multiple Introductions of Zika Virus into the United States. Nature 2017, 546, 401–405. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Saraf, S.; Gangavarapu, K.; Watts, A.; Tan, A.L.; Oidtman, R.J.; Ladner, J.T.; Oliveira, G.; Matteson, N.L.; Kraemer, M.U.G.; et al. Travel Surveillance and Genomics Uncover a Hidden Zika Outbreak during the Waning Epidemic. Cell 2019, 178, 1057–1071.e11. [Google Scholar] [CrossRef]
- Chitti, S.V.; Prasad, A.K.; Saxena, S.K. Emerging Zika Virus Disease: A Public Health Emergency of Global Concern. Virus Dis. 2016, 27, 211–214. [Google Scholar] [CrossRef]
- WHO. WHO Director-General Summarizes the Outcome of the Emergency Committee Regarding Clusters of Microcephaly and Guillain-Barré Syndrome. Available online: https://www.who.int/en/news-room/detail/01-02-2016-who-director-general-summarizes-the-outcome-of-the-emergency-committee-regarding-clusters-of-microcephaly-and-guillain-barré-syndrome (accessed on 1 April 2023).
- WHO. Fifth Meeting of the Emergency Committee under the International Health Regulations. Regarding Microcephaly, Other Neurological Disorders and Zika Virus. 2005. Available online: https://www.who.int/en/news-room/detail/18-11-2016-fifth-meeting-of-the-emergency-committee-under-the-international-health-regulations-(2005)-regarding-microcephaly-other-neurological-disorders-and-zika-virus (accessed on 1 April 2023).
- Faria, N.R.; da Silva Azevedo, R.D.S.; Kraemer, M.U.G.; Souza, R.; Cunha, M.S.; Hill, S.C.; Thézé, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika Virus in the Americas: Early Epidemiological and Genetic Findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.; Beau De Rochars, V.M.; El Badry, M.; Loeb, J.; Telisma, T.; Chavannes, S.; Anilis, G.; Cella, E.; Ciccozzi, M.; Rashid, M.; et al. Zika Virus Outbreak in Haiti in 2014: Molecular and Clinical Data. PLoS Negl. Trop. Dis. 2016, 10, e0004687. [Google Scholar] [CrossRef]
- Campos, T.D.L.; Durães-Carvalho, R.; Rezende, A.M.; de Carvalho, O.V.; Kohl, A.; Wallau, G.L.; Pena, L.J. Revisiting Key Entry Routes of Human Epidemic Arboviruses into the Mainland Americas through Large-Scale Phylogenomics. Int. J. Genom. 2018, 2018, 6941735. [Google Scholar] [CrossRef] [PubMed]
- Villabona-Arenas, C.J.; Hanage, W.P.; Tully, D.C. Phylogenetic Interpretation during Outbreaks Requires Caution. Nat. Microbiol. 2020, 5, 876–877. [Google Scholar] [CrossRef]
- Corman, V.M.; Rasche, A.; Baronti, C.; Aldabbagh, S.; Cadar, D.; Reusken, C.B.; Pas, S.D.; Goorhuis, A.; Schinkel, J.; Molenkamp, R.; et al. Assay Optimization for Molecular Detection of Zika Virus. Bull. World Health Organ. 2016, 94, 880–892. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.S.; St. John, A.L. Cross-Reactive Immunity Among Flaviviruses. Front. Immunol. 2020, 11, 334. [Google Scholar] [CrossRef]
- Cardoso, C.W.; Paploski, I.A.D.; Kikuti, M.; Rodrigues, M.S.; Silva, M.M.O.; Campos, G.S.; Sardi, S.I.; Kitron, U.; Reis, M.G.; Ribeiro, G.S. Outbreak of Exanthematous Illness Associated with Zika, Chikungunya, and Dengue Viruses, Salvador, Brazil. Emerg. Infect. Dis. 2015, 21, 2274–2276. [Google Scholar] [CrossRef]
- Silva, M.M.O.; Tauro, L.B.; Kikuti, M.; Anjos, R.O.; Santos, V.C.; Gonçalves, T.S.F.; Paploski, I.A.D.; Moreira, P.S.S.; Nascimento, L.C.J.; Campos, G.S.; et al. Concomitant Transmission of Dengue, Chikungunya, and Zika Viruses in Brazil: Clinical and Epidemiological Findings from Surveillance for Acute Febrile Illness. Clin. Infect. Dis. 2019, 69, 1353–1359. [Google Scholar] [CrossRef]
- de Moraes, L.; Cerqueira-Silva, T.; Nobrega, V.; Akrami, K.; Santos, L.A.; Orge, C.; Casais, P.; Cambui, L.; Rampazzo, R.d.C.P.; Trinta, K.S.; et al. A Clinical Scoring System to Predict Long-Term Arthralgia in Chikungunya Disease: A Cohort Study. PLoS Negl. Trop. Dis. 2020, 14, e0008467. [Google Scholar] [CrossRef]
- Balm, M.N.D.; Lee, C.K.; Lee, H.K.; Chiu, L.; Koay, E.S.C.; Tang, J.W. A Diagnostic Polymerase Chain Reaction Assay for Zika Virus. J. Med. Virol. 2012, 84, 1501–1505. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR Method for MinION and Illumina Sequencing of Zika and Other Virus Genomes Directly from Clinical Samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Black, A.; Moncla, L.H.; Laiton-Donato, K.; Potter, B.; Pardo, L.; Rico, A.; Tovar, C.; Rojas, D.P.; Longini, I.M.; Halloran, M.E.; et al. Genomic Epidemiology Supports Multiple Introductions and Cryptic Transmission of Zika Virus in Colombia. BMC Infect. Dis. 2019, 19, 963. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; De Pristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.-M.; et al. Genome Detective: An Automated System for Virus Identification from High-Throughput Sequencing Data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics. 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Baele, G.; Ayres, D.L.; Rambaut, A.; Suchard, M.A.; Lemey, P. High-Performance Computing in Bayesian Phylogenetics and Phylodynamics Using BEAGLE. In Evolutionary Genomics; Humana: New York, NY, USA, 2019; Volume 1910, pp. 691–722. [Google Scholar]
- Ayres, D.L.; Cummings, M.P.; Baele, G.; Darling, A.E.; Lewis, P.O.; Swofford, D.L.; Huelsenbeck, J.P.; Lemey, P.; Rambaut, A.; Suchard, M.A. BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2019, 68, 1052–1061. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Yang, Z. Maximum Likelihood Phylogenetic Estimation from DNA Sequences with Variable Rates over Sites: Approximate Methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef]
- Edwards, C.J.; Suchard, M.A.; Lemey, P.; Welch, J.J.; Barnes, I.; Fulton, T.L.; Barnett, R.; O’Connell, T.C.; Coxon, P.; Monaghan, N.; et al. Ancient Hybridization and an Irish Origin for the Modern Polar Bear Matriline. Curr. Biol. 2011, 21, 1251–1258. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A. Bayesian Random Local Clocks, or One Rate to Rule Them All. BMC Biol. 2010, 8, 114. [Google Scholar] [CrossRef]
- Minin, V.N.; Suchard, M.A. Fast, Accurate and Simulation-Free Stochastic Mapping. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3985–3995. [Google Scholar] [CrossRef] [PubMed]
- Minin, V.N.; Suchard, M.A. Counting Labeled Transitions in Continuous-Time Markov Models of Evolution. J. Math. Biol. 2007, 56, 391–412. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.; Ho, S.Y.W.; Drummond, A.J.; Suchard, M.A.; Pybus, O.G.; Rambaut, A. A Bayesian Phylogenetic Method to Estimate Unknown Sequence Ages. Mol. Biol. Evol. 2011, 28, 879–887. [Google Scholar] [CrossRef]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian Population Dynamics Inference: A Coalescent-Based Model for Multiple Loci. Mol. Biol. Evol. 2013, 30, 713–724. [Google Scholar] [CrossRef]
- Ferreira, M.A.R.; Suchard, M.A. Bayesian Analysis of Elapsed Times in Continuous-Time Markov Chains. Can. J. Stat. 2008, 36, 355–368. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Ho, Z.J.M.; Hapuarachchi, H.C.; Barkham, T.; Chow, A.; Ng, L.C.; Lee, J.M.V.; Leo, Y.S.; Prem, K.; Lim, Y.H.G.; de Sessions, P.F.; et al. Outbreak of Zika Virus Infection in Singapore: An Epidemiological, Entomological, Virological, and Clinical Analysis. Lancet Infect. Dis. 2017, 17, 813–821. [Google Scholar] [CrossRef]
- Massad, E.; Burattini, M.N.; Khan, K.; Struchiner, C.J.; Coutinho, F.A.B.; Wilder-Smith, A. On the Origin and Timing of Zika Virus Introduction in Brazil. Epidemiol. Infect. 2017, 145, 2303–2312. [Google Scholar] [CrossRef]
- Metsky, H.C.; Matranga, C.B.; Wohl, S.; Schaffner, S.F.; Freije, C.A.; Winnicki, S.M.; West, K.; Qu, J.; Baniecki, M.L.; Gladden-Young, A.; et al. Zika Virus Evolution and Spread in the Americas. Nature 2017, 546, 411–415. [Google Scholar] [CrossRef]
- Faria, N.R.; Sabino, E.C.; Nunes, M.R.T.; Alcantara, L.C.J.; Loman, N.J.; Pybus, O.G. Mobile Real-Time Surveillance of Zika Virus in Brazil. Genome Med. 2016, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Quick, J.; Claro, I.M.; Thézé, J.; de Jesus, J.G.; Giovanetti, M.; Kraemer, M.U.G.; Hill, S.C.; Black, A.; da Costa, A.C.; et al. Establishment and Cryptic Transmission of Zika Virus in Brazil and the Americas. Nature 2017, 546, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Iani, F.C.M.; Giovanetti, M.; Fonseca, V.; Souza, W.M.; Adelino, T.E.R.; Xavier, J.; Jesus, J.G.; Pereira, M.A.; Silva, M.V.F.; Costa, A.V.B.; et al. Epidemiology and Evolution of Zika Virus in Minas Gerais, Southeast Brazil. Infect. Genet. Evol. 2021, 91, 104785. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Pereira, L.A.; Adelino, T.É.R.; Fonseca, V.; Xavier, J.; de Araújo Fabri, A.; Slavov, S.N.; da Silva Lemos, P.; de Almeida Marques, W.; Kashima, S.; et al. A Retrospective Overview of Zika Virus Evolution in the Midwest of Brazil. Microbiol. Spectr. 2022, 10, e00155-22. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, M.; Faria, N.R.; Lourenço, J.; Goes de Jesus, J.; Xavier, J.; Claro, I.M.; Kraemer, M.U.G.; Fonseca, V.; Dellicour, S.; Thézé, J.; et al. Genomic and Epidemiological Surveillance of Zika Virus in the Amazon Region. Cell Rep. 2020, 30, 2275–2283.e7. [Google Scholar] [CrossRef]
| ID | Municipality | Collection Date | Cq Value | No. of Mapped Reads | Avg. Depth Coverage | Reference Covered |
|---|---|---|---|---|---|---|
| TRDP173 | Salvador | 6 May 2015 | 30.96 | 39,617 | 1207.66 | 94.03 |
| TRDP238 | Salvador | 18 May 2015 | 31.16 | 44,842 | 1258.69 | 90.22 |
| TRDP252 | Salvador | 19 May 2015 | 28.64 | 38,452 | 1197.17 | 88.76 |
| TRDP256 | Salvador | 19 May 2015 | 29.95 | 39,530 | 1170.28 | 89.10 |
| TRDP257 | Salvador | 19 May 2015 | 37.96 | 58,214 | 1372.15 | 83.82 |
| TRDP274 | Salvador | 20 May 2015 | 37.84 | 35,944 | 1116.41 | 84.62 |
| TRDP282 | Salvador | 21 May 2015 | 35.49 | 21,026 | 727.62 | 88.89 |
| TRDP300 | Salvador | 22 May 2015 | 30.03 | 19,670 | 678.25 | 81.89 |
| TRDP309 | Salvador | 26 May 2015 | 33.87 | 20,459 | 705.72 | 87.94 |
| TRDP317 | Salvador | 26 May 2015 | 33.60 | 20,414 | 704.03 | 87.99 |
| TRDP333 | Salvador | 27 May 2015 | 39.07 | 11,078 | 377.35 | 80.62 |
| TRDP433 | Salvador | 9 July 2015 | 27.63 | 60,626 | 1438.09 | 96.64 |
| ZK0110 | Campo Formoso | 8 April 2016 | 21.91 | 40,452 | 1301.48 | 97.18 |
| ZK0152 | Campo Formoso | 9 April 2016 | 34.94 | 17,820 | 562.33 | 84.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Moraes, L.; Portilho, M.M.; Vrancken, B.; Van den Broeck, F.; Santos, L.A.; Cucco, M.; Tauro, L.B.; Kikuti, M.; Silva, M.M.O.; Campos, G.S.; et al. Analyses of Early ZIKV Genomes Are Consistent with Viral Spread from Northeast Brazil to the Americas. Viruses 2023, 15, 1236. https://doi.org/10.3390/v15061236
de Moraes L, Portilho MM, Vrancken B, Van den Broeck F, Santos LA, Cucco M, Tauro LB, Kikuti M, Silva MMO, Campos GS, et al. Analyses of Early ZIKV Genomes Are Consistent with Viral Spread from Northeast Brazil to the Americas. Viruses. 2023; 15(6):1236. https://doi.org/10.3390/v15061236
Chicago/Turabian Stylede Moraes, Laise, Moyra M. Portilho, Bram Vrancken, Frederik Van den Broeck, Luciane Amorim Santos, Marina Cucco, Laura B. Tauro, Mariana Kikuti, Monaise M. O. Silva, Gúbio S. Campos, and et al. 2023. "Analyses of Early ZIKV Genomes Are Consistent with Viral Spread from Northeast Brazil to the Americas" Viruses 15, no. 6: 1236. https://doi.org/10.3390/v15061236
APA Stylede Moraes, L., Portilho, M. M., Vrancken, B., Van den Broeck, F., Santos, L. A., Cucco, M., Tauro, L. B., Kikuti, M., Silva, M. M. O., Campos, G. S., Reis, M. G., Barral, A., Barral-Netto, M., Boaventura, V. S., Vandamme, A.-M., Theys, K., Lemey, P., Ribeiro, G. S., & Khouri, R. (2023). Analyses of Early ZIKV Genomes Are Consistent with Viral Spread from Northeast Brazil to the Americas. Viruses, 15(6), 1236. https://doi.org/10.3390/v15061236






