Status and Developing Strategies for Neutralizing Monoclonal Antibody Therapy in the Omicron Era of COVID-19
Abstract
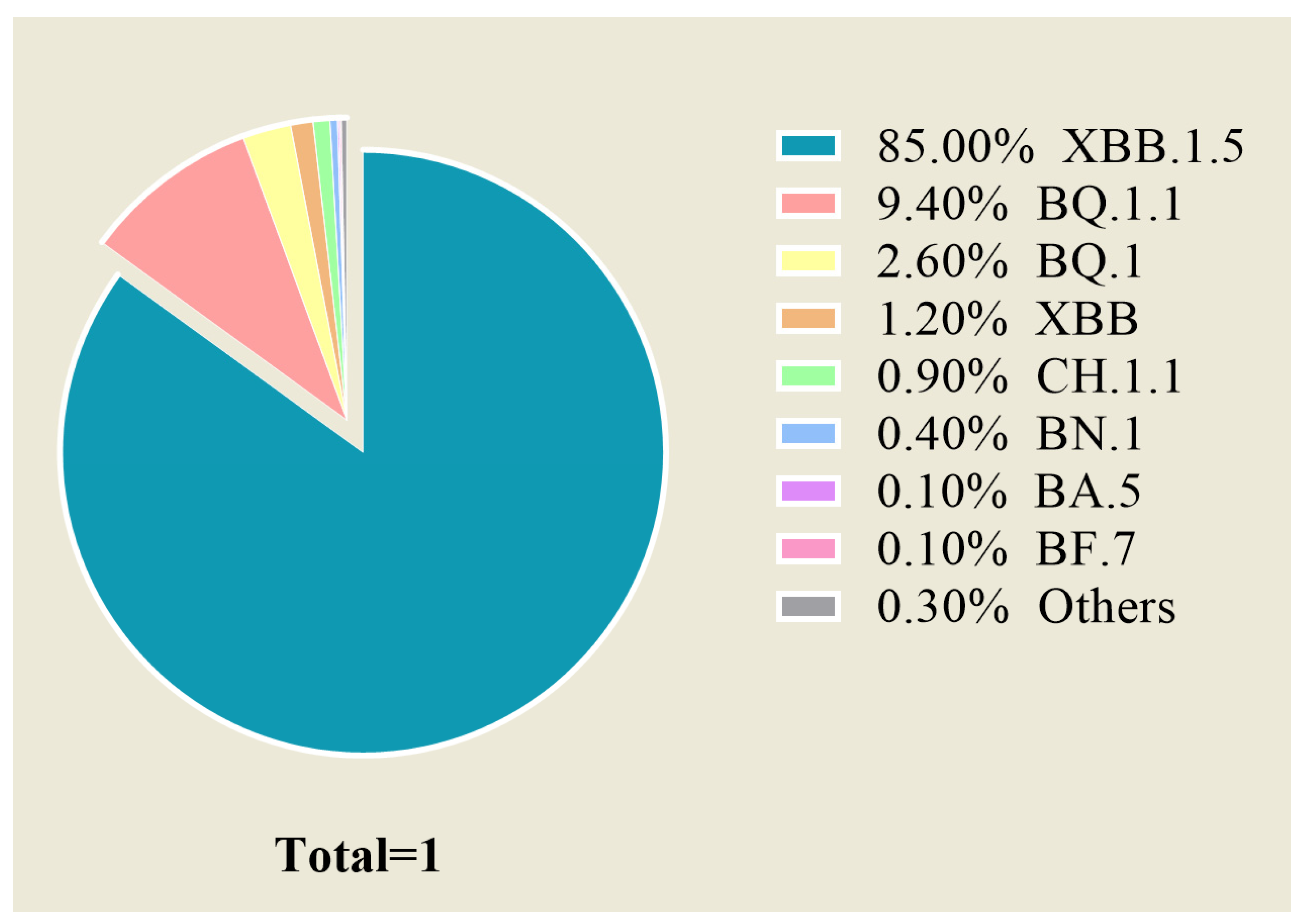
:1. Introduction
2. Main Text
2.1. Most Existing Therapeutic mAbs Targeting the RBD Have Lost the Ability to Neutralize the Current Omicron Variants
| Variants | Amino Acid Sequence of S protein RBD | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region of RBM | ||||||||||||||||||||||||||||||
| 339 | 346 | 356 | 368 | 371 | 373 | 375 | 376 | 405 | 408 | 417 | 440 | 444 | 445 | 446 | 452 | 455 | 460 | 477 | 478 | 484 | 486 | 490 | 493 | 496 | 498 | 501 | 505 | 519 | ||
| Alpha | B.1.1.7 | G | R | K | L | S | S | S | T | D | R | K | N | K | V | G | L | L | N | S | T | E | F | F | Q | G | Q | Y | Y | H |
| Beta | B.1.351 | G | R | K | L | S | S | S | T | D | R | N | N | K | V | G | L | L | N | S | T | K | F | F | Q | G | Q | Y | Y | H |
| Delta | B.1.617.2 | G | R | K | L | S | S | S | T | D | R | K | N | K | V | G | R | L | N | S | K | E | F | F | Q | G | Q | N | Y | H |
| Omicron | BA.1 | D | R | K | L | L | P | F | T | D | R | N | K | K | V | S | L | X | N | N | K | A | F | F | R | X | R | Y | H | X |
| BA.2 | D | T | K | L | S | S | S | T | D | R | K | N | K | V | G | L | L | N | S | T | E | F | F | Q | G | Q | N | Y | H | |
| BA.2.12.1 | D | R | K | L | F | P | F | A | N | S | N | K | K | V | G | Q | L | N | N | K | A | F | F | R | G | R | Y | H | H | |
| BA.4 | D | R | K | L | F | P | F | A | N | S | N | K | K | V | G | R | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BA.4.6 | D | T | K | L | F | P | F | A | N | S | K | K | K | V | G | R | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BA.5 | D | R | K | L | F | P | F | A | D | R | K | N | K | V | G | L | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BA.5.2.6 | D | T | K | L | F | P | F | A | N | S | N | K | K | V | G | R | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BE.1.1.1 | D | R | K | L | F | P | F | A | N | S | N | K | T | V | G | R | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BF.11 | D | T | K | L | F | P | F | A | N | S | N | K | K | V | G | R | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BF.7 | D | T | K | L | F | P | F | A | N | S | N | K | K | V | G | R | L | N | N | K | A | V | F | Q | G | R | Y | H | H | |
| BQ.1 | D | R | K | L | F | P | F | A | N | S | N | K | T | V | G | R | L | K | X | K | A | V | F | Q | G | R | Y | H | H | |
| BJ.1 | H | T | K | I | F | P | F | A | N | S | N | K | K | P | S | L | L | K | N | K | A | S | S | Q | G | R | Y | H | H | |
| BA.2.75.2 | H | T | K | L | F | P | F | A | N | S | N | K | K | V | S | L | L | K | N | K | A | S | F | Q | G | R | Y | H | H | |
| BM.4.1 | G | R | K | L | S | S | S | T | D | R | K | N | K | V | G | L | L | N | S | K | A | S | F | Q | G | R | Y | H | H | |
| BM.4.1.1 | H | T | K | L | F | P | F | A | D | R | K | N | K | V | G | L | L | N | N | K | A | S | F | Q | G | R | Y | H | H | |
| BN.1 | H | T | T | L | F | P | F | A | N | S | N | K | K | V | S | L | L | K | N | K | A | F | S | Q | G | R | Y | H | H | |
| CH.1.1.1 | H | T | K | L | F | P | F | A | N | S | N | K | T | V | S | R | L | K | N | K | A | S | F | Q | G | R | Y | H | H | |
| XBB.1 | H | T | K | I | F | P | F | A | N | S | N | K | K | P | S | L | L | K | N | K | A | S | S | Q | G | R | Y | H | H | |
| XBB.1.5 | Y | T | K | I | F | P | F | A | N | S | N | K | K | P | S | L | L | K | N | K | A | P | S | Q | G | R | Y | H | H | |
| XBB.1.9 | H | T | K | I | F | P | F | A | N | S | N | K | K | P | S | L | L | K | N | K | A | S | S | Q | G | R | Y | H | H | |
| SARS-CoV-2 Variants | Variations(+) in Therapeutic Epitopes or Binding Sites (VinTEBS) on S Protein | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N501Y | K417N | E484K | L452R | E484Q | T478K | Q493R | G446S | S371L; G496S | S477N; S373P; G339D; E484A; Q498R; S375F; N440K; Y505H | S371F; D405N; R408S | L452Q | R346T | F486V | N460Y; N334K | K444T | N460K | R346K; F490V; V483A | G339R | F486S | F490S | K356T | V445A; V445L | F486L | ||||
| Alpha | B.1.1.7 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| Beta | B.1.351 | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| Delta | B.1.617.2 | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| Omicron | BA.1 | + | + | − | − | − | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| BA.2 | BA.2 | + | + | − | − | - | + | + | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| BA.2.12.1 | + | + | − | − | - | + | + | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | |||
| BA.4 | BA.4 | + | + | − | + | - | + | − | − | −- | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| BA.4.6 | + | + | − | + | - | + | − | − | − | + | + | − | + | + | − | − | − | − | − | − | − | − | − | − | |||
| BA.5 | BA.5 | + | + | − | + | - | + | − | − | − | + | + | − | − | + | − | − | − | − | − | − | − | − | − | − | ||
| BA.5.2.6 | + | + | − | + | - | + | − | − | − | + | + | − | + | + | + | − | − | − | − | − | − | − | − | − | |||
| BE.1.1.1 | + | + | − | + | - | + | − | − | − | + | + | − | − | + | − | + | − | − | − | − | − | − | − | − | |||
| BF.11 | + | + | − | + | - | + | − | − | − | + | + | − | + | + | − | − | − | − | − | − | − | − | − | − | |||
| BF.7 | + | + | − | + | - | + | − | − | − | + | + | − | + | + | − | − | − | − | − | − | − | − | − | − | |||
| BQ.1 | + | + | − | + | - | + | − | − | − | + | + | − | − | + | − | + | + | − | − | − | − | − | − | − | |||
| BJ.1 | + | + | − | − | − | + | + | + | − | + | + | - | − | − | − | − | − | + | − | − | − | − | − | − | |||
| BA.2.75 | BA.2.75.2 | + | + | − | − | − | + | + | − | − | + | + | − | + | − | − | − | + | − | + | + | − | − | − | − | ||
| BM.4.1 | + | + | − | − | − | + | + | − | − | + | + | − | − | − | − | − | + | − | + | + | − | − | − | − | |||
| BM.4.1.1 | + | + | − | − | − | + | + | − | − | + | + | − | + | − | − | − | + | − | + | + | − | − | − | − | |||
| BN.1 | + | + | − | − | − | + | + | − | − | + | + | − | + | − | − | − | + | − | − | − | + | + | − | − | |||
| CH.1.1.1 | + | + | − | + | − | + | + | − | − | + | + | − | + | − | − | + | + | − | + | + | − | − | − | − | |||
| XBB | XBB.1 | + | + | − | − | − | + | − | − | − | + | + | − | + | − | − | − | + | − | + | + | + | − | + | − | ||
| XBB.1.5 | + | + | − | − | − | + | − | + | − | + | + | − | + | − | − | − | + | − | + | + | + | − | + | + | |||
| XBB.1.9 | + | + | − | − | − | + | − | + | − | + | + | − | + | − | − | − | + | − | + | + | + | − | + | − | |||
2.2. Strategies for Neutralizing mAbs Development Confronting the Viral Evasion of Omicron VOCs
2.2.1. Optimization of Epitope Selection for SARS-CoV-2
Optimizing the Selection of S Protein Epitopes
Monoclonal Antibodies That Do Not Directly Neutralize the Virus but Instead Target Human ACE2 Receptors
Block the Other Routes for Viral Entry in Addition to the RBD-ACE2
Take Full Advantage of Non-Neutralizing Antibodies That Target Conserved Epitopes in the S Protein
2.2.2. Enhance Antibody Potency
Cocktail and Bispecific Antibody
Nanobodies
IgM, IgA, and Polymeric IgG
2.2.3. Optimizing Immunization Regimens, such as Antigen Components, Immunization Routes, and Adjuvants
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sheervalilou, R.; Shirvaliloo, M.; Dadashzadeh, N.; Shirvalilou, S.; Shahraki, O.; Pilehvar-Soltanahmadi, Y.; Ghaznavi, H.; Khoei, S.; Nazarlou, Z. COVID-19 under spotlight: A close look at the origin, transmission, diagnosis, and treatment of the 2019-nCoV disease. J. Cell. Physiol. 2020, 235, 8873–8924. [Google Scholar] [CrossRef] [PubMed]
- Gentile, I.; Schiano Moriello, N. COVID-19 prophylaxis in immunosuppressed patients: Beyond vaccination. PLoS Med. 2022, 19, e1003917. [Google Scholar] [CrossRef] [PubMed]
- Pashaei, M.; Rezaei, N. Immunotherapy for SARS-CoV-2: Potential opportunities. Expert Opin. Biol. Ther. 2020, 20, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Shanmugaraj, B.; Siriwattananon, K.; Wangkanont, K.; Phoolcharoen, W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac. J. Allergy Immunol. 2020, 38, 10–18. [Google Scholar] [CrossRef]
- Weiner, L.M.; Surana, R.; Wang, S. Monoclonal antibodies: Versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 2010, 10, 317–327. [Google Scholar] [CrossRef]
- Bajic, G.; Degn, S.E.; Thiel, S.; Andersen, G.R. Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J. 2015, 34, 2735–2757. [Google Scholar] [CrossRef]
- Wang, X.Y.; Wang, B.; Wen, Y.M. From therapeutic antibodies to immune complex vaccines. NPJ Vaccines 2019, 4, 2. [Google Scholar] [CrossRef]
- Teoh, K.T.; Siu, Y.L.; Chan, W.L.; Schlüter, M.A.; Liu, C.J.; Peiris, J.S.; Bruzzone, R.; Margolis, B.; Nal, B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell 2010, 21, 3838–3852. [Google Scholar] [CrossRef]
- Fu, Y.Z.; Wang, S.Y.; Zheng, Z.Q.; Yi, H.; Li, W.W.; Xu, Z.S.; Wang, Y.Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell. Mol. Immunol. 2021, 18, 613–620. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Peiris, J.S.; Guan, Y.; Yuen, K.Y. Severe acute respiratory syndrome. Nat. Med. 2004, 10 (Suppl. 12), S88–S97. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.Y.; Ong, H.K.; Yeap, S.K.; Ho, K.L.; Tan, W.S. Recent Advances in the Vaccine Development Against Middle East Respiratory Syndrome-Coronavirus. Front. Microbiol. 2019, 10, 1781. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Jia, W.; Wang, P.; Zhang, S.; Shi, X.; Wang, X.; Zhang, L. Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg. Microbes Infect. 2019, 8, 841–856. [Google Scholar] [CrossRef]
- Coughlin, M.M.; Prabhakar, B.S. Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: Target, mechanism of action, and therapeutic potential. Rev. Med. Virol. 2012, 22, 2–17. [Google Scholar] [CrossRef]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e1021. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Qing, E.; Kicmal, T.; Kumar, B.; Hawkins, G.M.; Timm, E.; Perlman, S.; Gallagher, T. Dynamics of SARS-CoV-2 Spike Proteins in Cell Entry: Control Elements in the Amino-Terminal Domains. mBio 2021, 12, e0159021. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, T.; Zhang, J.; Shao, B.; Gong, H.; Wang, Y.; He, X.; Liu, S.; Liu, T.Y. Exploring the Regulatory Function of the N-terminal Domain of SARS-CoV-2 Spike Protein through Molecular Dynamics Simulation. Adv. Theory Simul. 2021, 4, 2100152. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Gonzalez-Rojas, Y.; Juarez, E.; Crespo Casal, M.; Moya, J.; Rodrigues Falci, D.; Sarkis, E.; Solis, J.; Zheng, H.; Scott, N.; et al. Effect of Sotrovimab on Hospitalization or Death Among High-risk Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2022, 327, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.M.; Deveau, C.; Upadhyaya, H. Bebtelovimab: Considerations for global access to treatments during a rapidly evolving pandemic. Lancet. Infect. Dis. 2022, 22, 1531. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef]
- Gottlieb, R.L.; Nirula, A.; Chen, P.; Boscia, J.; Heller, B.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA 2021, 325, 632–644. [Google Scholar] [CrossRef]
- Levin, M.J.; Ustianowski, A.; De Wit, S.; Launay, O.; Avila, M.; Templeton, A.; Yuan, Y.; Seegobin, S.; Ellery, A.; Levinson, D.J.; et al. Intramuscular AZD7442 (Tixagevimab-Cilgavimab) for Prevention of COVID-19. N. Engl. J. Med. 2022, 386, 2188–2200. [Google Scholar] [CrossRef]
- FDA. Coronavirus (COVID-19)|Drugs. Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed on 23 February 2023).
- World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 26 November 2021).
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 26 November 2021).
- GISAID. Tracking of hCoV 19 Variants. Available online: https://gisaidorg/hcov19-variants/ (accessed on 23 February 2023).
- Telenti, A.; Arvin, A.; Corey, L.; Corti, D.; Diamond, M.S.; García-Sastre, A.; Garry, R.F.; Holmes, E.C.; Pang, P.S.; Virgin, H.W. After the pandemic: Perspectives on the future trajectory of COVID-19. Nature 2021, 596, 495–504. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, P.; Wang, N.; Wang, L.; Fan, K.; Zhu, Q.; Wang, K.; Chen, R.; Feng, R.; Jia, Z.; et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871.e813. [Google Scholar] [CrossRef]
- Peter, A.S.; Grüner, E.; Socher, E.; Fraedrich, K.; Richel, E.; Mueller-Schmucker, S.; Cordsmeier, A.; Ensser, A.; Sticht, H.; Überla, K. Characterization of SARS-CoV-2 Escape Mutants to a Pair of Neutralizing Antibodies Targeting the RBD and the NTD. Int. J. Mol. Sci. 2022, 23, 8177. [Google Scholar] [CrossRef]
- Syed, A.M.; Ciling, A.; Taha, T.Y.; Chen, I.P.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Kumar, G.R.; Suryawanshi, R.; Silva, I.; et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. Proc. Natl. Acad. Sci. USA 2022, 119, e2200592119. [Google Scholar] [CrossRef] [PubMed]
- Moulana, A.; Dupic, T.; Phillips, A.M.; Chang, J.; Roffler, A.A.; Greaney, A.J.; Starr, T.N.; Bloom, J.D.; Desai, M.M. The landscape of antibody binding affinity in SARS-CoV-2 Omicron BA.1 evolution. eLife 2023, 12, 83442. [Google Scholar] [CrossRef] [PubMed]
- Kupferschmidt, K. Where did ’weird’ Omicron come from? Science 2021, 374, 1179. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wu, L.; Zheng, A.; Xie, Y.; He, Q.; Rong, X.; Han, P.; Du, P.; Han, P.; Zhang, Z.; et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3. Immunity 2022, 55, 1501–1514.e1503. [Google Scholar] [CrossRef]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; di Iulio, J.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef]
- Hong, Q.; Han, W.; Li, J.; Xu, S.; Wang, Y.; Xu, C.; Li, Z.; Wang, Y.; Zhang, C.; Huang, Z.; et al. Molecular basis of receptor binding and antibody neutralization of Omicron. Nature 2022, 604, 546–552. [Google Scholar] [CrossRef]
- Pinto, D.; Park, Y.J.; Beltramello, M.; Walls, A.C.; Tortorici, M.A.; Bianchi, S.; Jaconi, S.; Culap, K.; Zatta, F.; De Marco, A.; et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583, 290–295. [Google Scholar] [CrossRef]
- Wang, Z.; Muecksch, F.; Cho, A.; Gaebler, C.; Hoffmann, H.H.; Ramos, V.; Zong, S.; Cipolla, M.; Johnson, B.; Schmidt, F.; et al. Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins. Immunity 2022, 55, 998–1012.e1018. [Google Scholar] [CrossRef]
- Casadevall, A.; Focosi, D. SARS-CoV-2 variants resistant to monoclonal antibodies in immunocompromised patients constitute a public health concern. J. Clin. Investig. 2023, 133, jci168603. [Google Scholar] [CrossRef]
- NCBI. Bold Text Represents Amino Acid Mutations. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=taxid:2697049 (accessed on 23 February 2023).
- NCBI. SARS-CoV-2 Variants Overview. Available online: https://www.ncbi.nlm.nih.gov/activ (accessed on 23 February 2023).
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Jette, C.A.; Cohen, A.A.; Gnanapragasam, P.N.P.; Muecksch, F.; Lee, Y.E.; Huey-Tubman, K.E.; Schmidt, F.; Hatziioannou, T.; Bieniasz, P.D.; Nussenzweig, M.C.; et al. Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep. 2021, 36, 109760. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host. Microb. 2021, 29, 463–476.e466. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Cohen, A.A.; Park, M.; Hung, A.F.; Keeffe, J.R.; Gnanapragasam, P.N.P.; Lee, Y.E.; Gao, H.; Kakutani, L.M.; Wu, Z.; et al. Neutralizing monoclonal antibodies elicited by mosaic RBD nanoparticles bind conserved sarbecovirus epitopes. Immunity 2022, 55, 2419–2435.e2410. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Czudnochowski, N.; Liu, Z.; Zatta, F.; Park, Y.J.; Addetia, A.; Pinto, D.; Beltramello, M.; Hernandez, P.; Greaney, A.J.; et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 2021, 597, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, Y.; Hu, Y.; Chang, F.; Wu, Q.; Yang, J.; Chen, J.; Teng, S.; Zhang, J.; He, R.; et al. Monoclonal antibodies constructed from COVID-19 convalescent memory B cells exhibit potent binding activity to MERS-CoV spike S2 subunit and other human coronaviruses. Front. Immunol. 2022, 13, 1056272. [Google Scholar] [CrossRef] [PubMed]
- Errico, J.M.; Adams, L.J.; Fremont, D.H. Antibody-mediated immunity to SARS-CoV-2 spike. Adv. Immunol. 2022, 154, 1–69. [Google Scholar] [CrossRef]
- Zhou, P.; Song, G.; He, W.-t.; Beutler, N.; Tse, L.V.; Martinez, D.R.; Schäfer, A.; Anzanello, F.; Yong, P.; Peng, L.; et al. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause severe disease. bioRxiv 2022. [Google Scholar] [CrossRef]
- Crowley, A.R.; Natarajan, H.; Hederman, A.P.; Bobak, C.A.; Weiner, J.A.; Wieland-Alter, W.; Lee, J.; Bloch, E.M.; Tobian, A.A.R.; Redd, A.D.; et al. Boosting of cross-reactive antibodies to endemic coronaviruses by SARS-CoV-2 infection but not vaccination with stabilized spike. eLife 2022, 11, 75228. [Google Scholar] [CrossRef]
- Piepenbrink, M.S.; Park, J.G.; Deshpande, A.; Loos, A.; Ye, C.; Basu, M.; Sarkar, S.; Khalil, A.M.; Chauvin, D.; Woo, J.; et al. Potent universal beta-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody. PLoS Pathog. 2022, 18, e1010691. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shi, R.; Zhang, Y.; Duan, X.; Li, L.; Zhang, J.; Wang, F.; Zhang, R.; Shen, H.; Wang, Y.; et al. A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants. Nat. Commun. 2021, 12, 5000. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, Y.; Zhou, Z.; Zhang, Z.; Xiao, X.; Liu, Z.; Chen, A.; Dong, X.; Tian, F.; Chen, S.; et al. Genome-wide CRISPR activation screen identifies candidate receptors for SARS-CoV-2 entry. Sci. China Life Sci. 2022, 65, 701–717. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, Y.; Zhou, Z.; Zhang, Z.; Xiao, X.; Liu, Z.; Chen, A.; Dong, X.; Tian, F.; Chen, S.; et al. Genome-wide CRISPR activation screen identifies novel receptors for SARS-CoV-2 entry. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; He, J.; Jiang, X.; Zhang, J.; Shen, G.; et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 2022, 32, 24–37. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Song, Z.; Zhang, S.; Nanda, A.; Li, G. CD147: A novel modulator of inflammatory and immune disorders. Curr. Med. Chem. 2014, 21, 2138–2145. [Google Scholar] [CrossRef]
- Geng, J.; Chen, L.; Yuan, Y.; Wang, K.; Wang, Y.; Qin, C.; Wu, G.; Chen, R.; Zhang, Z.; Wei, D.; et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target. Ther. 2021, 6, 347. [Google Scholar] [CrossRef]
- Tian, J.; Yuan, X.; Xiao, J.; Zhong, Q.; Yang, C.; Liu, B.; Cai, Y.; Lu, Z.; Wang, J.; Wang, Y.; et al. Clinical characteristics and risk factors associated with COVID-19 disease severity in patients with cancer in Wuhan, China: A multicentre, retrospective, cohort study. Lancet. Oncol. 2020, 21, 893–903. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Qin, C.; Huo, F.; Liang, X.; Yang, X.; Zhang, K.; Lin, P.; Liu, J.; Feng, Z.; et al. CD147 contributes to SARS-CoV-2-induced pulmonary fibrosis. Signal Transduct. Target. Ther. 2022, 7, 382. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Chen, L.; Zheng, Z.H.; Sun, X.X.; Geng, J.J.; Chen, R.; Wang, K. Meplazumab in hospitalized adults with severe COVID-19 (DEFLECT): A multicenter, seamless phase 2/3, randomized, third-party double-blind clinical trial. Signal Transduct. Target. Ther. 2023, 8, 46. [Google Scholar] [CrossRef]
- Badeti, S.; Jiang, Q.; Naghizadeh, A.; Tseng, H.C.; Bushkin, Y.; Marras, S.A.E.; Nisa, A.; Tyagi, S.; Chen, F.; Romanienko, P.; et al. Development of a novel human CD147 knock-in NSG mouse model to test SARS-CoV-2 viral infection. Cell Biosci. 2022, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- VanBlargan, L.A.; Errico, J.M.; Halfmann, P.J.; Zost, S.J.; Crowe, J.E., Jr.; Purcell, L.A.; Kawaoka, Y.; Corti, D.; Fremont, D.H.; Diamond, M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022, 28, 490–495. [Google Scholar] [CrossRef]
- Lim, S.A.; Gramespacher, J.A.; Pance, K.; Rettko, N.J.; Solomon, P.; Jin, J.; Lui, I.; Elledge, S.K.; Liu, J.; Bracken, C.J.; et al. Bispecific VH/Fab antibodies targeting neutralizing and non-neutralizing Spike epitopes demonstrate enhanced potency against SARS-CoV-2. mAbs 2021, 13, 1893426. [Google Scholar] [CrossRef] [PubMed]
- Weidenbacher, P.A.B.; Waltari, E.; de los Rios Kobara, I.; Bell, B.N.; Morris, M.K.; Cheng, Y.-C.; Hanson, C.; Pak, J.E.; Kim, P.S. Converting non-neutralizing SARS-CoV-2 antibodies into broad-spectrum inhibitors. Nat. Chem. Biol. 2022, 18, 1270–1276. [Google Scholar] [CrossRef]
- Glasgow, A.; Glasgow, J.; Limonta, D.; Solomon, P.; Lui, I.; Zhang, Y.; Nix, M.A.; Rettko, N.J.; Zha, S.; Yamin, R.; et al. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 28046–28055. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Yurina, V.; Adianingsih, O.R. Predicting epitopes for vaccine development using bioinformatics tools. Ther. Adv. Vaccines Immunother. 2022, 10, 25151355221100218. [Google Scholar] [CrossRef] [PubMed]
- Gupta, Y.; Savytskyi, O.V.; Coban, M.; Venugopal, A.; Pleqi, V.; Weber, C.A.; Chitale, R.; Durvasula, R.; Hopkins, C.; Kempaiah, P.; et al. Protein structure-based in-silico approaches to drug discovery: Guide to COVID-19 therapeutics. Mol. Asp. Med. 2023, 91, 101151. [Google Scholar] [CrossRef]
- Gopal, R.; Fitzpatrick, E.; Pentakota, N.; Jayaraman, A.; Tharakaraman, K.; Capila, I. Optimizing Antibody Affinity and Developability Using a Framework-CDR Shuffling Approach-Application to an Anti-SARS-CoV-2 Antibody. Viruses 2022, 14, 2694. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- De Gasparo, R.; Pedotti, M.; Simonelli, L.; Nickl, P.; Muecksch, F.; Cassaniti, I.; Percivalle, E.; Lorenzi, J.C.C.; Mazzola, F.; Magrì, D.; et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature 2021, 593, 424–428. [Google Scholar] [CrossRef]
- Li, C.; Zhan, W.; Yang, Z.; Tu, C.; Hu, G.; Zhang, X.; Song, W.; Du, S.; Zhu, Y.; Huang, K.; et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell 2022, 185, 1389–1401.e1318. [Google Scholar] [CrossRef]
- Yuan, M.; Chen, X.; Zhu, Y.; Dong, X.; Liu, Y.; Qian, Z.; Ye, L.; Liu, P. A Bispecific Antibody Targeting RBD and S2 Potently Neutralizes SARS-CoV-2 Omicron and Other Variants of Concern. J. Virol. 2022, 96, e0077522. [Google Scholar] [CrossRef]
- Saied, A.A.; Metwally, A.A.; Alobo, M.; Shah, J.; Sharun, K.; Dhama, K. Bovine-derived antibodies and camelid-derived nanobodies as biotherapeutic weapons against SARS-CoV-2 and its variants: A review article. Int. J. Surg. 2022, 98, 106233. [Google Scholar] [CrossRef]
- Salvador, J.P.; Vilaplana, L.; Marco, M.P. Nanobody: Outstanding features for diagnostic and therapeutic applications. Anal. Bioanal. Chem. 2019, 411, 1703–1713. [Google Scholar] [CrossRef]
- Dormeshkin, D.; Shapira, M.; Dubovik, S.; Kavaleuski, A.; Katsin, M.; Migas, A.; Meleshko, A.; Semyonov, S. Isolation of an escape-resistant SARS-CoV-2 neutralizing nanobody from a novel synthetic nanobody library. Front. Immunol. 2022, 13, 965446. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; da Fonseca Rezende, E.M.J.; Fleming, B.D.; Renn, A.; Chen, C.Z.; Hu, X.; Xu, M.; Gorshkov, K.; Hanson, Q.; Zheng, W.; et al. A humanized nanobody phage display library yields potent binders of SARS CoV-2 spike. PLoS ONE 2022, 17, e0272364. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Nambulli, S.; Xiao, Z.; Liu, H.; Sang, Z.; Duprex, W.P.; Schneidman-Duhovny, D.; Zhang, C.; Shi, Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 2020, 370, 1479–1484. [Google Scholar] [CrossRef]
- Sun, D.; Sang, Z.; Kim, Y.J.; Xiang, Y.; Cohen, T.; Belford, A.K.; Huet, A.; Conway, J.F.; Sun, J.; Taylor, D.J.; et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat. Commun. 2021, 12, 4676. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Huang, B.; Jia, Z.; Wang, B.; Gallolu Kankanamalage, S.; Titong, A.; Liu, Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microbes Infect. 2020, 9, 1034–1036. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.I.; Morrison, S.L. Recombinant polymeric IgG: An approach to engineering more potent antibodies. Bio/Technol. Nat. Publ. 1994, 12, 683–688. [Google Scholar] [CrossRef]
- Ku, Z.; Xie, X.; Hinton, P.R.; Liu, X.; Ye, X.; Muruato, A.E.; Ng, D.C.; Biswas, S.; Zou, J.; Liu, Y.; et al. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature 2021, 595, 718–723. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Li, N.; Wang, Y.; Zhu, Q.; Chu, H.; Wu, W.; Tan, Y.; Yu, F.; Su, X.D.; et al. Structural insights into immunoglobulin M. Science 2020, 367, 1014–1017. [Google Scholar] [CrossRef]
- Callegari, I.; Schneider, M.; Berloffa, G.; Mühlethaler, T.; Holdermann, S.; Galli, E.; Roloff, T.; Boss, R.; Infanti, L.; Khanna, N.; et al. Potent neutralization by monoclonal human IgM against SARS-CoV-2 is impaired by class switch. EMBO Rep. 2022, 23, e53956. [Google Scholar] [CrossRef]
- Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Viant, C.; Gaebler, C.; Cipolla, M.; Hoffmann, H.H.; Oliveira, T.Y.; Oren, D.A.; et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021, 13, abf1555. [Google Scholar] [CrossRef]
- Havervall, S.; Marking, U.; Svensson, J.; Greilert-Norin, N.; Bacchus, P.; Nilsson, P.; Hober, S.; Gordon, M.; Blom, K.; Klingström, J.; et al. Anti-Spike Mucosal IgA Protection against SARS-CoV-2 Omicron Infection. N. Engl. J. Med. 2022, 387, 1333–1336. [Google Scholar] [CrossRef] [PubMed]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, C.; Yin, L.; Sun, J.; Wang, W.; Li, H.; Zhang, Z.; Chen, S.; Liu, B.; Liu, Z.; et al. Intranasal booster using an Omicron vaccine confers broad mucosal and systemic immunity against SARS-CoV-2 variants. Signal Transduct. Target. Ther. 2023, 8, 167. [Google Scholar] [CrossRef]
- Zou, J.; Li, L.; Zheng, P.; Liang, W.; Hu, S.; Zhou, S.; Wang, Y.; Zhao, J.; Yuan, D.; Liu, L.; et al. Ultrapotent neutralizing antibodies against SARS-CoV-2 with a high degree of mutation resistance. J. Clin. Investig. 2022, 132, jci154987. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Rakhra, K.; Abraham, W.; Wang, C.; Moynihan, K.D.; Li, N.; Donahue, N.; Baldeon, A.D.; Irvine, D.J. Exploiting albumin as a mucosal vaccine chaperone for robust generation of lung-resident memory T cells. Sci. Immunol. 2021, 6, abd8003. [Google Scholar] [CrossRef]
- Mao, T.; Israelow, B.; Peña-Hernández, M.A.; Suberi, A.; Zhou, L.; Luyten, S.; Reschke, M.; Dong, H.; Homer, R.J.; Saltzman, W.M.; et al. Unadjuvanted intranasal spike vaccine elicits protective mucosal immunity against sarbecoviruses. Science 2022, 378, eabo2523. [Google Scholar] [CrossRef]
- Cai, J.P.; Luo, C.; Wang, K.; Cao, H.; Chen, L.L.; Zhang, X.; Han, Y.; Yin, F.; Zhang, A.J.; Chu, H.; et al. Intranasal Boosting with Spike Fc-RBD of Wild-Type SARS-CoV-2 Induces Neutralizing Antibodies against Omicron Subvariants and Reduces Viral Load in the Nasal Turbinate of Mice. Viruses 2023, 15, 687. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Liu, J.; Fang, E.; Liu, X.; Peng, Q.; Zhang, Z.; Li, M.; Liu, X.; Wu, X.; et al. Combining intramuscular and intranasal homologous prime-boost with a chimpanzee adenovirus-based COVID-19 vaccine elicits potent humoral and cellular immune responses in mice. Emerg. Microbes Infect. 2022, 11, 1890–1899. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Ravetch, J.V. Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef]
- Du, L.; Zhao, G.; Chan, C.C.; Sun, S.; Chen, M.; Liu, Z.; Guo, H.; He, Y.; Zhou, Y.; Zheng, B.J.; et al. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology 2009, 393, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Arunachalam, P.S.; O’Hagan, D.T. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021, 20, 454–475. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, J.; Wang, X.; Xu, W.; Teng, Z.; Chen, H.; Chen, M.; Zhang, G.; Wang, Y.; Huang, J.; et al. A pan-sarbecovirus vaccine based on RBD of SARS-CoV-2 original strain elicits potent neutralizing antibodies against XBB in non-human primates. Proc. Natl. Acad. Sci. USA 2023, 120, e2221713120. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Z.; Shen, C.; Peng, J. Status and Developing Strategies for Neutralizing Monoclonal Antibody Therapy in the Omicron Era of COVID-19. Viruses 2023, 15, 1297. https://doi.org/10.3390/v15061297
Ren Z, Shen C, Peng J. Status and Developing Strategies for Neutralizing Monoclonal Antibody Therapy in the Omicron Era of COVID-19. Viruses. 2023; 15(6):1297. https://doi.org/10.3390/v15061297
Chicago/Turabian StyleRen, Zuning, Chenguang Shen, and Jie Peng. 2023. "Status and Developing Strategies for Neutralizing Monoclonal Antibody Therapy in the Omicron Era of COVID-19" Viruses 15, no. 6: 1297. https://doi.org/10.3390/v15061297
APA StyleRen, Z., Shen, C., & Peng, J. (2023). Status and Developing Strategies for Neutralizing Monoclonal Antibody Therapy in the Omicron Era of COVID-19. Viruses, 15(6), 1297. https://doi.org/10.3390/v15061297





