Gut Bacterial Diversity of Field and Laboratory-Reared Aedes albopictus Populations of Rio de Janeiro, Brazil
Abstract
1. Introduction
2. Materials and Methods
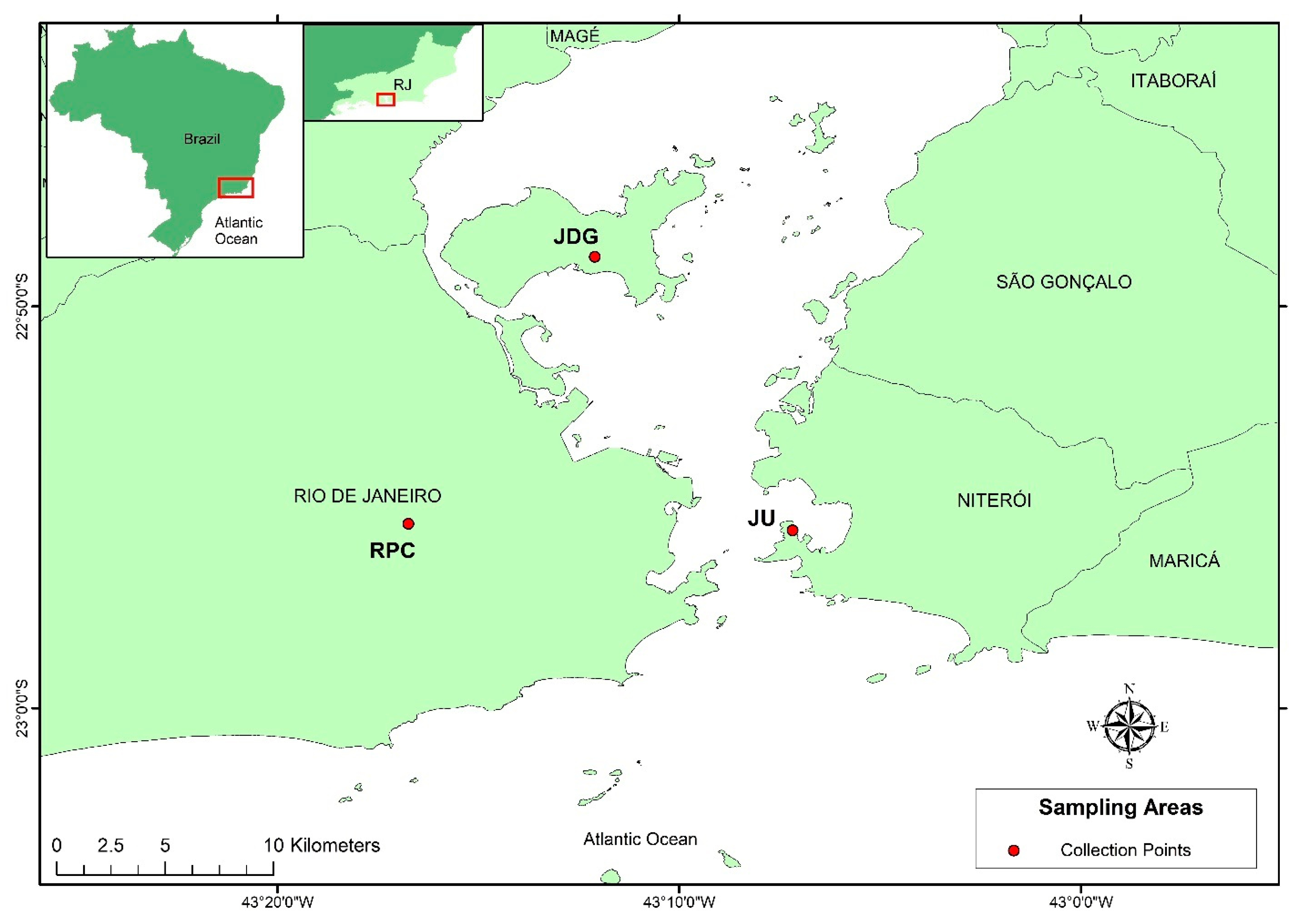
2.1. Study Areas
2.2. Adult Collection and Identification
2.3. Egg Collection and Mosquito Rearing
2.4. Gut Processing and DNA Extraction
2.5. Sequencing of the V3–V4 Region of the 16s rRNA Gene
2.6. Bioinformatic Analysis
2.7. Diversity Analysis
2.8. Taxonomic Composition, Differential Abundance Analysis and Core Microbiota
2.9. ZIKV Infection
3. Results
3.1. Sequencing Data
3.2. Microbiota Diversity
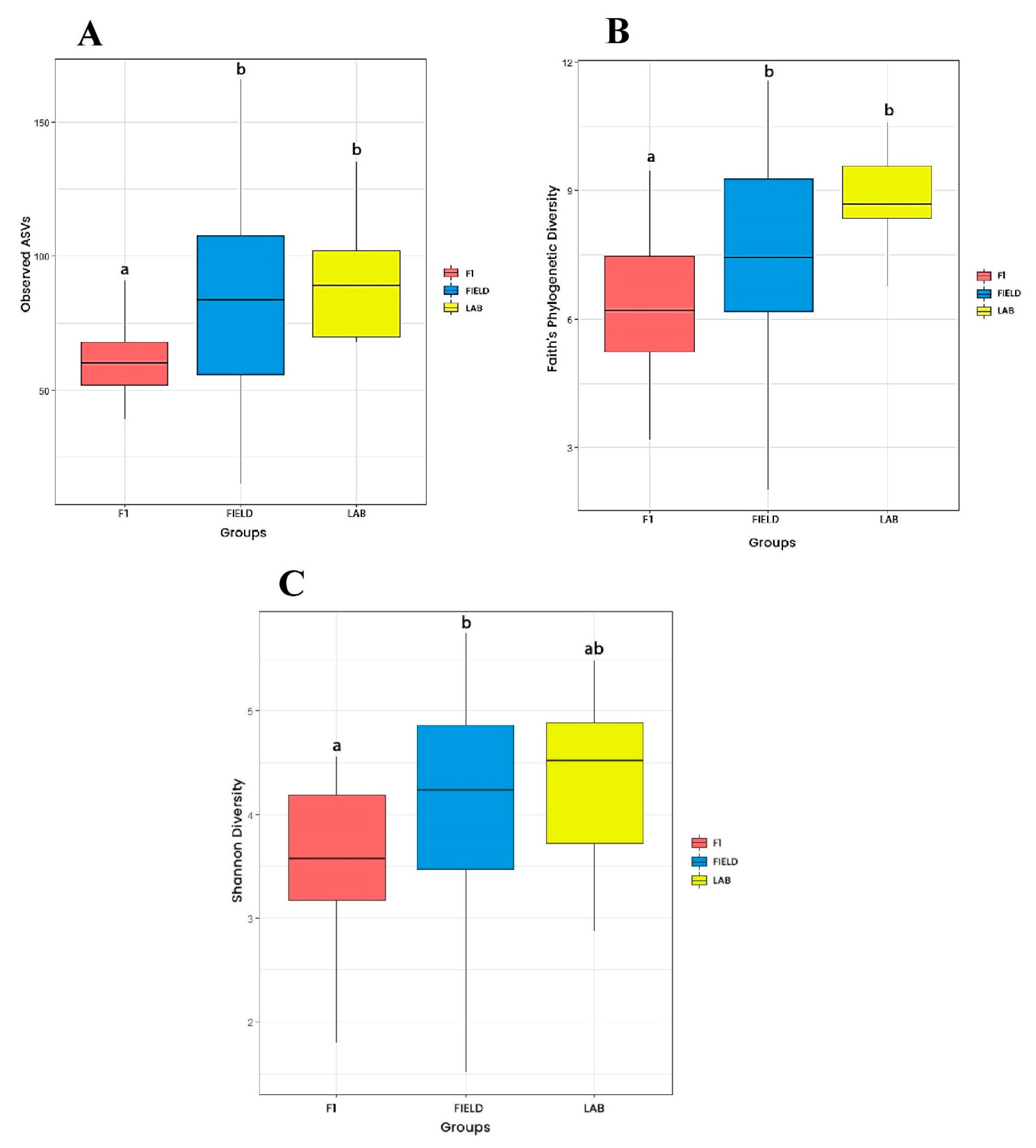
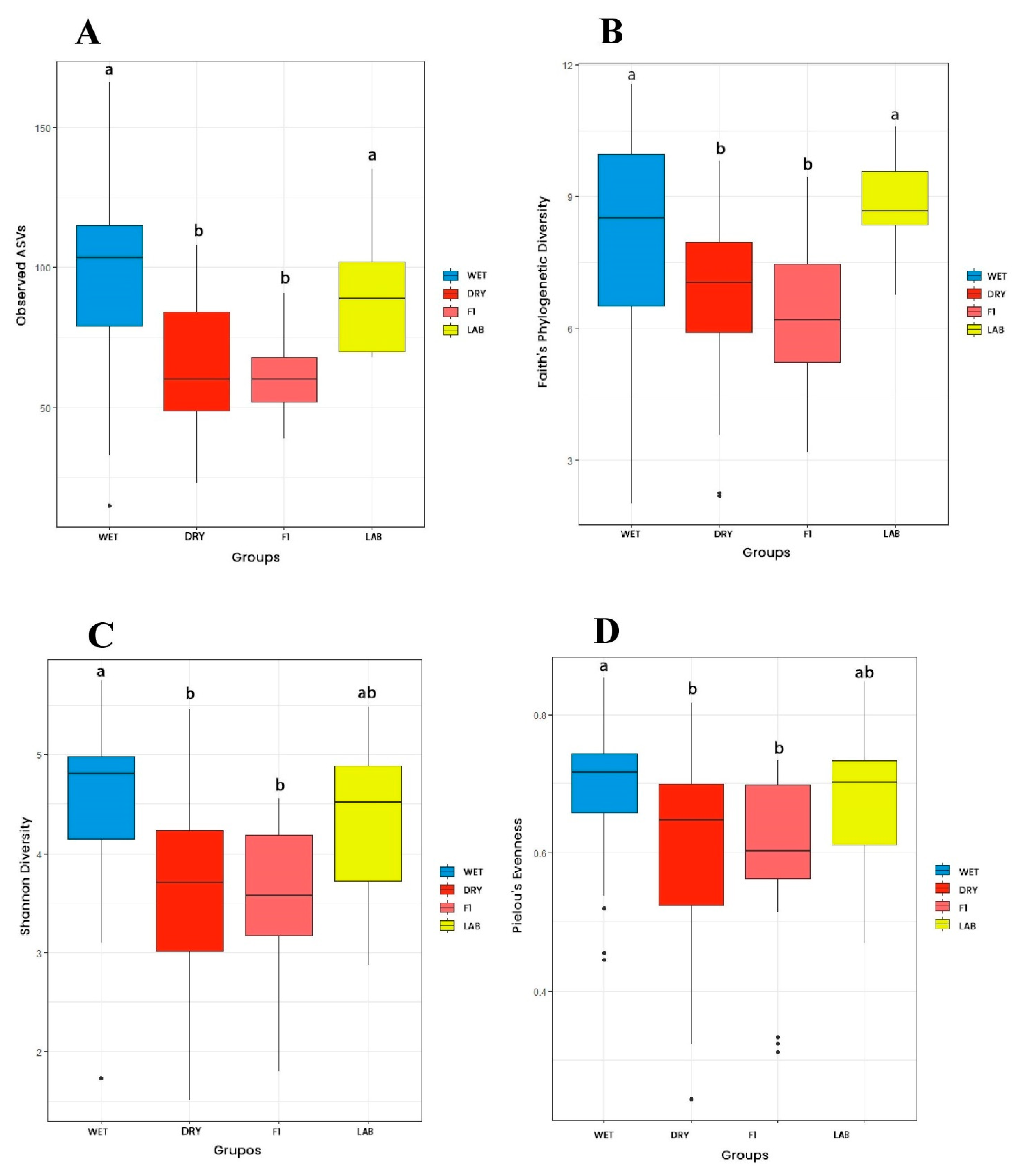
3.2.1. Alpha Diversity
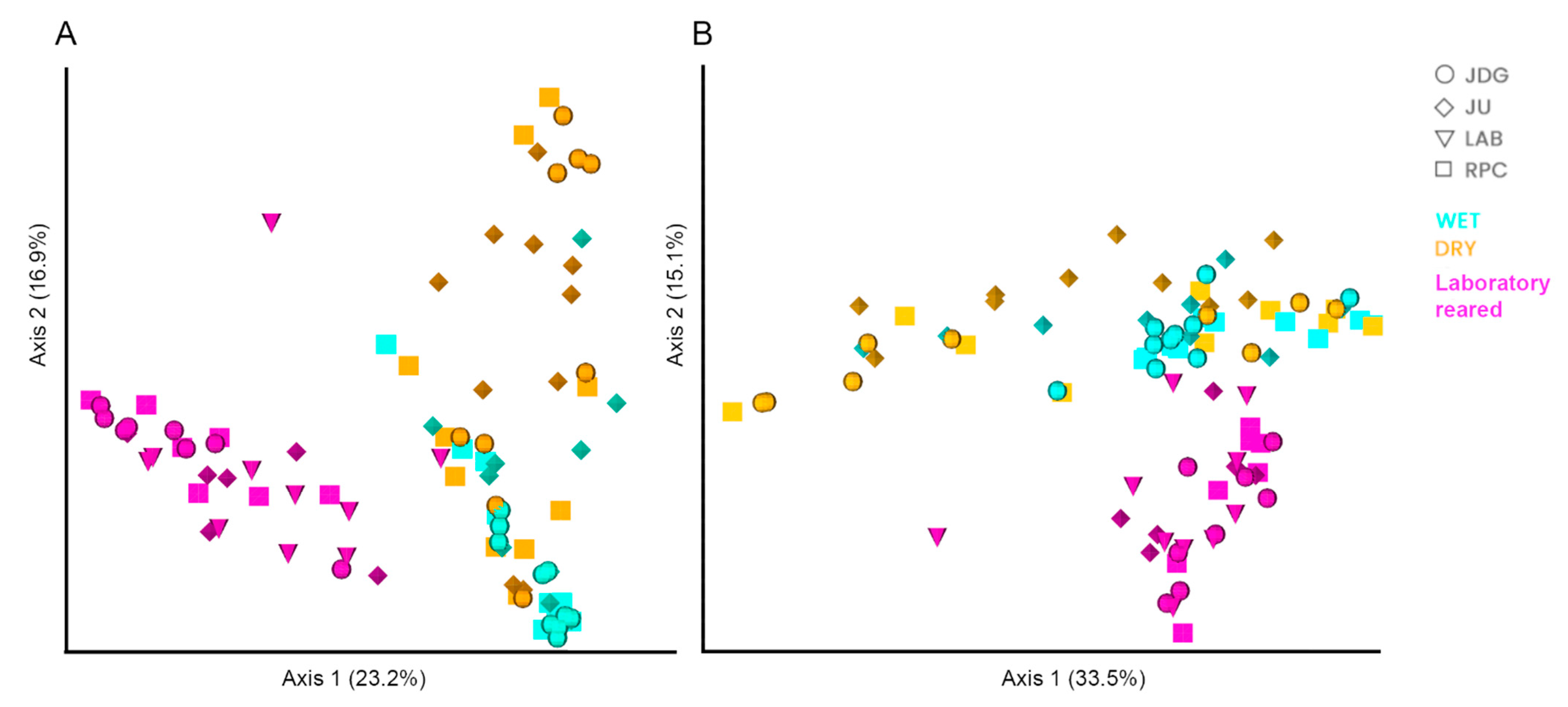
3.2.2. Beta Diversity
3.3. Aedes albopictus Microbiota Taxonomic Composition
3.4. Aedes albopictus Core Microbiota
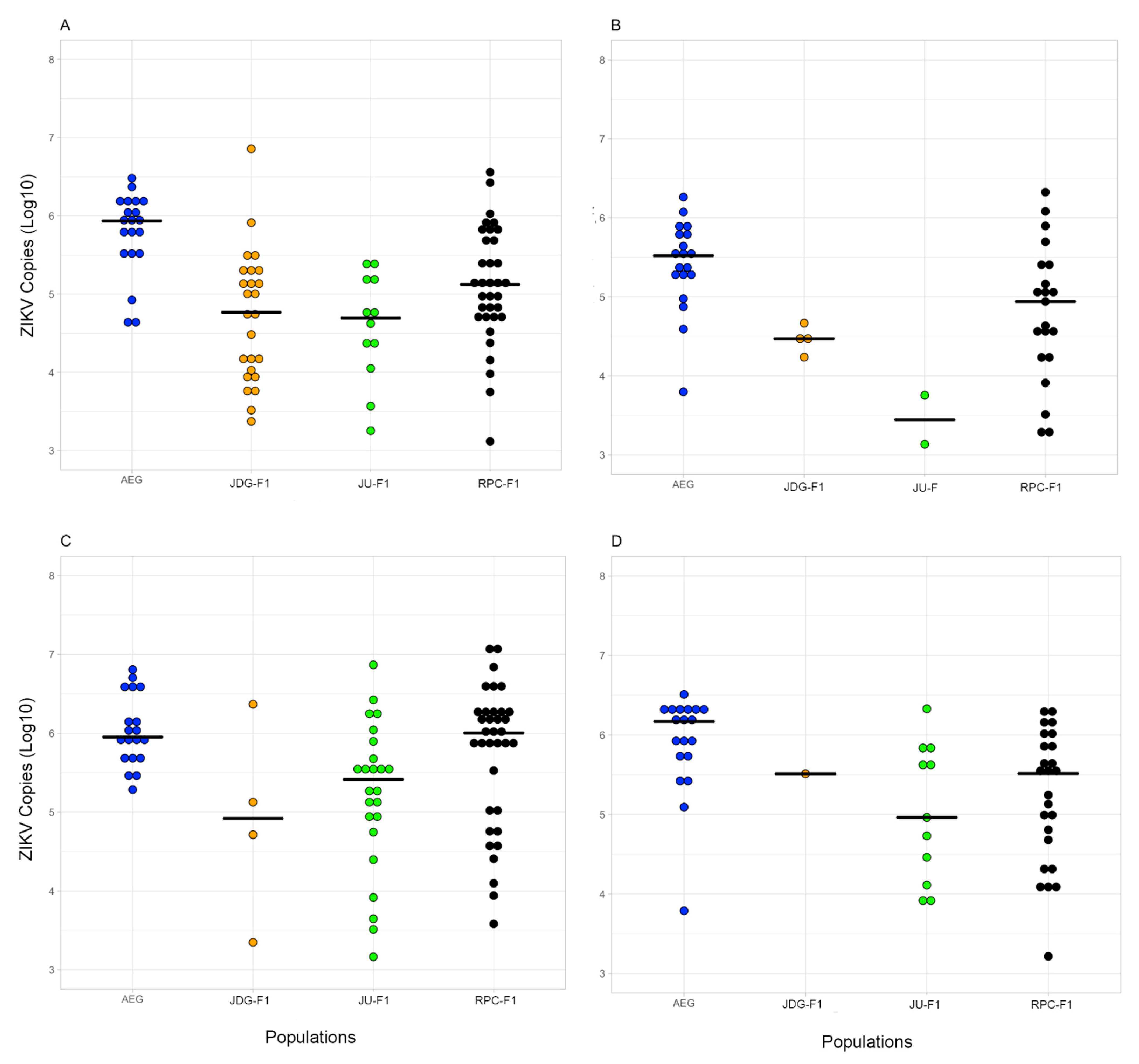
3.5. Vector Competence to ZIKV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harbach, R.E. Mosquito Taxonomic Inventory. Available online: https://mosquito-taxonomic-inventory.myspecies.info (accessed on 6 September 2022).
- Weaver, S.C.; Reisen, W.K. Present and Future Arboviral Threats. Antivir. Res. 2010, 85, 328–345. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-Y.; Lun, Z.-R.; James, A.A.; Chen, X.-G. Dengue Fever in Mainland China. Am. J. Trop. Med. Hyg. 2010, 83, 664. [Google Scholar] [CrossRef]
- Lazzarini, L.; Barzon, L.; Foglia, F.; Manfrin, V.; Pacenti, M.; Pavan, G.; Rassu, M.; Capelli, G.; Montarsi, F.; Martini, S.; et al. First Autochthonous Dengue Outbreak in Italy, August 2020. Eurosurveillance 2020, 25, 2001606. [Google Scholar] [CrossRef] [PubMed]
- Monge, S.; García-Ortúzar, V.; López Hernández, B.; Lopaz Pérez, M.Á.; Delacour-Estrella, S.; Sánchez-Seco, M.P.P.; Fernández Martinez, B.; García San Miguel, L.; García-Fulgueiras, A.; Sierra Moros, M.J.J. Characterization of the First Autochthonous Dengue Outbreak in Spain (August–September 2018). Acta Trop. 2020, 205, 105402. [Google Scholar] [CrossRef]
- Pagès, F.; Peyrefitte, C.N.; Mve, M.T.; Jarjaval, F.; Brisse, S.; Iteman, I.; Gravier, P.; Nkoghe, D.; Grandadam, M. Aedes albopictus Mosquito: The Main Vector of the 2007 Chikungunya Outbreak in Gabon. PLoS ONE 2009, 4, e4691. [Google Scholar] [CrossRef]
- Pereira-Dos-Santos, T.; Roiz, D.; Lourenço-De-Oliveira, R.; Paupy, C. A Systematic Review: Is Aedes albopictus an Efficient Bridge Vector for Zoonotic Arboviruses? Pathogens 2020, 9, 266. [Google Scholar] [CrossRef]
- Bonizzoni, M.; Gasperi, G.; Chen, X.; James, A.A. The Invasive Mosquito Species Aedes albopictus: Current Knowledge and Future Perspectives. Trends Parasitol. 2013, 29, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-de-Lima, V.H.; Câmara, D.C.P.; Honório, N.A.; Lima-Camara, T.N. The Asian Tiger Mosquito in Brazil: Observations on Biology and Ecological Interactions since Its First Detection in 1986. Acta Trop. 2020, 205, 105386. [Google Scholar] [CrossRef]
- Kamgang, B.; Nchoutpouen, E.; Simard, F.; Paupy, C. Notes on the Blood-Feeding Behavior of Aedes albopictus (Diptera: Culicidae) in Cameroon. Parasites Vectors 2012, 5, 57. [Google Scholar] [CrossRef]
- Pereira dos Santos, T.; Roiz, D.; Santos de Abreu, F.V.; Luz, S.L.B.; Santalucia, M.; Jiolle, D.; Santos Neves, M.S.A.; Simard, F.; Lourenço-de-Oliveira, R.; Paupy, C. Potential of Aedes albopictus as a Bridge Vector for Enzootic Pathogens at the Urban-Forest Interface in Brazil. Emerg. Microbes Infect. 2018, 7, 1–8. [Google Scholar] [CrossRef]
- Medley, K.A.; Westby, K.M.; Jenkins, D.G. Rapid Local Adaptation to Northern Winters in the Invasive Asian Tiger Mosquito Aedes albopictus: A Moving Target. J. Appl. Ecol. 2019, 56, 2518–2527. [Google Scholar] [CrossRef]
- Lambrechts, L.; Scott, T.W.; Gubler, D.J. Consequences of the Expanding Global Distribution of Aedes albopictus for Dengue Virus Transmission. PLoS Negl. Trop. Dis. 2010, 4, e646. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.S.; O’connor, O.; Bersot, M.I.L.; Girault, D.; Dokunengo, M.R.; Pocquet, N.; Dupont-Rouzeyrol, M.; Lourenço-De-oliveira, R. Vector Competence of Aedes aegypti, Aedes albopictus and Culex quinquefasciatus from Brazil and New Caledonia for Three Zika Virus Lineages. Pathogens 2020, 9, 575. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, J.M.T.; Araújo, R.G.P.; Melo, F.F.; Gonçalves, C.M.; Chaves, B.A.; Silva, B.M.; Silva, L.D.; Brandão, S.T.; Secundino, N.F.C.; Norris, D.E.; et al. Aedes (Stegomyia) Albopictus’ Dynamics Influenced by Spatiotemporal Characteristics in a Brazilian Dengue-Endemic Risk City. Acta Trop. 2016, 164, 431–437. [Google Scholar] [CrossRef]
- Ferreira-De-Lima, V.H.; Lima-Camara, T.N. Natural Vertical Transmission of Dengue Virus in Aedes aegypti and Aedes albopictus: A Systematic Review. Parasit. Vectors 2018, 11, 77. [Google Scholar] [CrossRef]
- Cansado-Utrilla, C.; Zhao, S.Y.; Mccall, P.J.; Coon, K.L.; Hughes, G.L. The Microbiome and Mosquito Vectorial Capacity: Rich Potential for Discovery and Translation. Microbiome 2021, 9, 111. [Google Scholar] [CrossRef]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti Toll Pathway Controls Dengue Virus Infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef]
- Dong, Y.; Manfredini, F.; Dimopoulos, G. Implication of the Mosquito Midgut Microbiota in the Defense against Malaria Parasites. PLoS Pathog. 2009, 5, e1000423. [Google Scholar] [CrossRef]
- Cirimotich, C.M.; Dong, Y.; Clayton, A.M.; Sandiford, S.L.; Souza-Neto, J.A.; Mulenga, M.; Dimopoulos, G. Natural Microbe-Mediated Refractoriness to Plasmodium Infection in Anopheles gambiae. Science 2011, 332, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Apte-Deshpande, A.D.; Paingankar, M.S.; Gokhale, M.D.; Deobagkar, D.N. Serratia Odorifera Mediated Enhancement in Susceptibility of Aedes aegypti for Chikungunya Virus. Indian J. Med. Res. 2014, 139, 762–768. [Google Scholar] [PubMed]
- Caragata, E.P.; Tikhe, C.V.; Dimopoulos, G. Curious Entanglements: Interactions between Mosquitoes, Their Microbiota, and Arboviruses. Curr. Opin. Virol. 2019, 37, 26–36. [Google Scholar] [CrossRef]
- Saraiva, R.G.; Dimopoulos, G. Bacterial Natural Products in the Fight against Mosquito-Transmitted Tropical Diseases. Nat. Prod. Rep. 2020, 37, 338–354. [Google Scholar] [CrossRef]
- Charan, S.S.; Pawar, K.D. Comparative Analysis of Midgut Bacterial Communities of Aedes aegypti Mosquito Strains Varying in Vector Competence to Dengue Virus. Parasitol. Res. 2013, 112, 2627–2637. [Google Scholar] [CrossRef] [PubMed]
- Charan, S.S.; Pawar, K.D.; Gavhale, S.D.; Tikhe, C.V.; Charan, N.S.; Angel, B.; Joshi, V.; Patole, M.S.; Shouche, Y.S. Comparative Analysis of Midgut Bacterial Communities in Three Aedine Mosquito Species from Dengue-Endemic and Non-Endemic Areas of Rajasthan, India. Med. Vet. Entomol. 2016, 30, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Cortés, A.; Damania, A.; Granada, Y.; Zuluaga, S.; Mejia, R.; Triana-Chavez, O. Association of Midgut Bacteria and Their Metabolic Pathways with Zika Infection and Insecticide Resistance in Colombian Aedes aegypti Populations. Viruses 2022, 14, 2197. [Google Scholar] [CrossRef]
- Coon, K.L.; Vogel, K.J.; Brown, M.R.; Strand, M.R. Mosquitoes Rely on Their Gut Microbiota for Development. Mol. Ecol. 2014, 23, 2727–2739. [Google Scholar] [CrossRef]
- Gaio, A.D.O.; Gusmão, D.S.; Santos, A.V.; Berbert-Molina, M.A.; Pimenta, P.F.P.; Lemos, F.J.A. Contribution of Midgut Bacteria to Blood Digestion and Egg Production in Aedes aegypti (Diptera: Culicidae) (L.). Parasit. Vectors 2011, 4, 105. [Google Scholar] [CrossRef]
- Guégan, M.; Tran Van, V.; Martin, E.; Minard, G.; Tran, F.H.; Fel, B.; Hay, A.E.; Simon, L.; Barakat, M.; Potier, P.; et al. Who Is Eating Fructose within the Aedes albopictus Gut Microbiota? Environ. Microbiol. 2020, 22, 1193–1206. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, F.H.; Gendrin, M.; Wyer, C.A.S.; Christophides, G.K. Microbiota-Induced Peritrophic Matrix Regulates Midgut Homeostasis and Prevents Systemic Infection of Malaria Vector Mosquitoes. PLoS Pathog. 2017, 13, e1006391. [Google Scholar] [CrossRef]
- Bascuñán, P.; Niño-Garcia, J.P.; Galeano-Castañeda, Y.; Serre, D.; Correa, M.M. Factors Shaping the Gut Bacterial Community Assembly in Two Main Colombian Malaria Vectors. Microbiome 2018, 6, 146. [Google Scholar] [CrossRef]
- Muturi, E.J.; Lagos-Kutz, D.; Dunlap, C.; Ramirez, J.L.; Rooney, A.P.; Hartman, G.L.; Fields, C.J.; Rendon, G.; Kim, C.H. Mosquito Microbiota Cluster by Host Sampling Location. Parasites Vectors 2018, 11, 468. [Google Scholar] [CrossRef]
- Bogale, H.N.; Cannon, M.V.; Keita, K.; Camara, D.; Barry, Y.; Keita, M.; Coulibaly, D.; Kone, A.K.; Doumbo, O.K.; Thera, M.A.; et al. Relative Contributions of Various Endogenous and Exogenous Factors to the Mosquito Microbiota. Parasites Vectors 2020, 13, 619. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, D.W.; Ramos, M.M.; Payne, K.C.; Giordano, B.V.; Caragata, E.P. Collection Time, Location, and Mosquito Species Have Distinct Impacts on the Mosquito Microbiota. Front. Trop. Dis. 2022, 3, 896289. [Google Scholar] [CrossRef]
- Buck, M.; Nilsson, L.K.J.; Brunius, C.; Dabiré, R.K.; Hopkins, R.; Terenius, O. Bacterial Associations Reveal Spatial Population Dynamics in Anopheles gambiae Mosquitoes. Sci. Rep. 2016, 6, 22806. [Google Scholar] [CrossRef] [PubMed]
- Coon, K.L.; Brown, M.R.; Strand, M.R. Mosquitoes Host Communities of Bacteria That Are Essential for Development but Vary Greatly between Local Habitats. Mol. Ecol. 2016, 25, 5806–5826. [Google Scholar] [CrossRef]
- Lindh, J.M.; Borg-Karlson, A.-K.; Faye, I. Transstadial and Horizontal Transfer of Bacteria within a Colony of Anopheles gambiae (Diptera: Culicidae) and Oviposition Response to Bacteria-Containing Water. Acta Trop. 2008, 107, 242–250. [Google Scholar] [CrossRef]
- Dillon, R.J.; Dillon, V.M. The Gut Bacteria of Insects: Nonpathogenic Interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef]
- Fridman, S.; Izhaki, I.; Gerchman, Y.; Halpern, M. Bacterial Communities in Floral Nectar. Environ. Microbiol. Rep. 2012, 4, 97–104. [Google Scholar] [CrossRef]
- Bassene, H.; Hadji, E.; Niang, A.; Fenollar, F.; Doucoure, S.; Faye, O.; Raoult, D.; Sokhna, C.; Mediannikov, O. Role of Plants in the Transmission of Asaia sp., which Potentially Inhibit the Plasmodium Sporogenic Cycle in Anopheles Mosquitoes. Sci. Rep. 2020, 10, 7144. [Google Scholar] [CrossRef]
- Honório, N.A.; Silva, W.D.C.; Leite, P.J.; Gonçalves, J.M.; Lounibos, L.P.; Lourenço-de-Oliveira, R. Dispersal of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in an Urban Endemic Dengue Area in the State of Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2003, 98, 191–198. [Google Scholar] [CrossRef]
- De Oliveira, S.; Villela, D.A.M.; Dias, F.B.S.; Moreira, L.A.; Maciel de Freitas, R. How Does Competition among Wild Type Mosquitoes Influence the Performance of Aedes aegypti and Dissemination of Wolbachia Pipientis? PLoS Negl. Trop. Dis. 2017, 11, e0005947. [Google Scholar] [CrossRef] [PubMed]
- INMET. Instituto Nacional de Meteorologia. Tabela de Dados Das Estações. Available online: https://tempo.inmet.gov.br/CondicoesRegistradas (accessed on 1 September 2019).
- Consoli, R.A.; Oliveira, R.L.D. Principais Mosquitos de Importância Sanitária No Brasil; Editora Fiocruz: Rio de Janeiro, RJ, Brazil, 1994. [Google Scholar]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol. 2019, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Illumina. 16S Metagenomic Sequencing Library; Illumina: San Diego, CA, USA, 2013; pp. 1–28. [Google Scholar]
- Illumina. BaseSpace Sequence Hub; Illumina: San Diego, CA, USA, 2017; pp. 1–6. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; Desantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. MicroDecon: A Highly Accurate Read-Subtraction Tool for the Post-Sequencing Removal of Contamination in Metabarcoding Studies. Environ. DNA 2019, 1, 14–25. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E. Qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. Available online: https://github.com/jbisanz/qiime2R (accessed on 1 July 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Heal. Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Guégan, M.; Zouache, K.; Démichel, C.; Minard, G.; Tran Van, V.; Potier, P.; Mavingui, P.; Valiente Moro, C. The Mosquito Holobiont: Fresh Insight into Mosquito-Microbiota Interactions. Microbiome 2018, 6, 49. [Google Scholar] [CrossRef]
- Rutledge, L.C.; Ward, R.A.; Gould, D.J. Studies on the Feeding Response of Mosquitoes to Nutritive Solutions in a New Membrane Feeder. Mosq. News 1964, 24, 407–419. [Google Scholar]
- Faria, N.R.; Azevedo, R.d.S.d.S.; Kraemer, M.U.G.; Souza, R.; Cunha, M.S.; Hill, S.C.; Thézé, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika Virus in the Americas: Early Epidemiological and Genetic Findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef]
- Fernandes, R.S.; Campos, S.S.; Ferreira-de-Brito, A.; Miranda, R.M.D.; Barbosa da Silva, K.A.; de Castro, M.G.; Raphael, L.M.S.; Brasil, P.; Failloux, A.-B.; Bonaldo, M.C.; et al. Culex quinquefasciatus from Rio de Janeiro Is Not Competent to Transmit the Local Zika Virus. PLoS Negl. Trop. Dis. 2016, 10, e0004993. [Google Scholar] [CrossRef]
- Ferreira-De-Brito, A.; Ribeiro, I.P.; De Miranda, R.M.; Fernandes, R.S.; Campos, S.S.; Da Silva, K.A.B.; De Castro, M.G.; Bonaldo, M.C.; Brasil, P.; Lourenço-De-Oliveira, R. First Detection of Natural Infection of Aedes aegypti with Zika Virus in Brazil and throughout South America. Mem. Inst. Oswaldo Cruz 2016, 111, 655–658. [Google Scholar] [CrossRef]
- Bonaldo, M.C.; Ribeiro, I.P.; Lima, N.S.; dos Santos, A.A.C.; Menezes, L.S.R.; da Cruz, S.O.D.; de Mello, I.S.; Furtado, N.D.; de Moura, E.E.; Damasceno, L.; et al. Isolation of Infective Zika Virus from Urine and Saliva of Patients in Brazil. PLoS Negl. Trop. Dis. 2016, 10, e0004816. [Google Scholar] [CrossRef]
- Minard, G.; Tran, F.H.; Dubost, A.; Tran-Van, V.; Mavingui, P.; Valiente Moro, C. Pyrosequencing 16S RRNA Genes of Bacteria Associated with Wild Tiger Mosquito Aedes albopictus: A Pilot Study. Front. Cell. Infect. Microbiol. 2014, 4, 59. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Wang, Y.; Li, S.; Sun, X.; Lu, X.; Rajaofera, M.J.N.; Lu, Y.; Kang, L.; Zheng, A.; Zou, Z.; et al. Comparative Analysis of the Gut Microbiota of Adult Mosquitoes from Eight Locations in Hainan, China. Front. Cell. Infect. Microbiol. 2020, 10, 596750. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Yek, S.H.; Wilson, R.F.; Rahman, S. Characterization of the Aedes albopictus (Diptera: Culicidae) Holobiome: Bacterial Composition across Land Use Type and Mosquito Sex in Malaysia. Acta Trop. 2020, 212, 105683. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, M.C.I.; Seabourn, P.S.; Rollins, R.L.; Yoneishi, N.M. Mosquito Microbiome Diversity Varies Along a Landscape–Scale Moisture Gradient. Microb. Ecol. 2021, 84, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Tawidian, P.; Coon, K.L.; Jumpponen, A.; Cohnstaedt, L.W.; Michel, K. Host-Environment Interplay Shapes Fungal Diversity in Mosquitoes. Msphere 2021, 6, e00646-21. [Google Scholar] [CrossRef]
- Wang, Y.; Gilbreath, T.M.; Kukutla, P.; Yan, G.; Xu, J. Dynamic Gut Microbiome across Life History of the Malaria Mosquito Anopheles gambiae in Kenya. PLoS ONE 2011, 6, e24767. [Google Scholar] [CrossRef]
- Minard, G.; Mavingui, P.; Moro, C.V. Diversity and Function of Bacterial Microbiota in the Mosquito Holobiont. Parasites Vectors 2013, 6, 146. [Google Scholar] [CrossRef]
- Scolari, F.; Casiraghi, M.; Bonizzoni, M. Aedes spp. and Their Microbiota: A Review. Front. Microbiol. 2019, 10, 2036. [Google Scholar] [CrossRef]
- Yadav, K.K.; Datta, S.; Naglot, A.; Bora, A.; Hmuaka, V.; Bhagyawant, S.; Gogoi, H.K.; Veer, V.; Raju, P.S. Diversity of Cultivable Midgut Microbiota at Different Stages of the Asian Tiger Mosquito, Aedes albopictus from Tezpur, India. PLoS ONE 2016, 11, e0167409. [Google Scholar] [CrossRef]
- Lin, D.; Zheng, X.; Sanogo, B.; Ding, T.; Sun, X.; Wu, Z. Bacterial Composition of Midgut and Entire Body of Laboratory Colonies of Aedes aegypti and Aedes albopictus from Southern China. Parasites Vectors 2021, 14, 586. [Google Scholar] [CrossRef]
- Bennett, K.L.; Gómez-Martínez, C.; Chin, Y.; Saltonstall, K.; McMillan, W.O.; Rovira, J.R.; Loaiza, J.R. Dynamics and Diversity of Bacteria Associated with the Disease Vectors Aedes aegypti and Aedes albopictus. Sci. Rep. 2019, 9, 12160. [Google Scholar] [CrossRef] [PubMed]
- Pereira, T.N.; Carvalho, F.D.; Rugani, J.N.; de Carvalho, V.R.; Jarusevicius, J.; Souza-Neto, J.A.; Moreira, L.A. Mayaro Virus: The Potential Role of Microbiota and Wolbachia. Pathogens 2021, 10, 525. [Google Scholar] [CrossRef] [PubMed]
- Seabourn, P.; Spafford, H.; Yoneishi, N.; Medeirosid, M. The Aedes albopictus (Diptera: Culicidae) Microbiome Varies Spatially and with Ascogregarine Infection. PLoS Negl. Trop. Dis. 2020, 14, e0008615. [Google Scholar] [CrossRef] [PubMed]
- Novakova, E.; Woodhams, D.C.; Rodríguez-Ruano, S.M.; Brucker, R.M.; Leff, J.W.; Maharaj, A.; Amir, A.; Knight, R.; Scott, J. Mosquito Microbiome Dynamics, a Background for Prevalence and Seasonality of West Nile Virus. Front. Microbiol. 2017, 8, 526. [Google Scholar] [CrossRef]
- Arévalo-Cortés, A.; Mejia-Jaramillo, A.M.; Granada, Y.; Coatsworth, H.; Lowenberger, C.; Triana-Chavez, O. The Midgut Microbiota of Colombian Aedes aegypti Populations with Different Levels of Resistance to the Insecticide Lambda-Cyhalothrin. Insects 2020, 11, 584. [Google Scholar] [CrossRef]
- Akorli, J.; Gendrin, M.; Pels, N.A.P.; Yeboah-manu, D. Seasonality and Locality Affect the Diversity of Anopheles gambiae and Anopheles coluzzii Midgut Microbiota from Ghana. PLoS ONE 2016, 11, e0157529. [Google Scholar] [CrossRef]
- Thongsripong, P.; Chandler, J.A.; Green, A.B.; Kittayapong, P.; Wilcox, B.A.; Kapan, D.D.; Bennett, S.N. Mosquito Vector-Associated Microbiota: Metabarcoding Bacteria and Eukaryotic Symbionts across Habitat Types in Thailand Endemic for Dengue and Other Arthropod-Borne Diseases. Ecol. Evol. 2017, 8, 1352–1368. [Google Scholar] [CrossRef]
- Li, Y.; Kamara, F.; Zhou, G.; Puthiyakunnon, S.; Li, C.; Liu, Y.; Zhou, Y.; Yao, L.; Yan, G.; Chen, X.-G. Urbanization Increases Aedes albopictus Larval Habitats and Accelerates Mosquito Development and Survivorship. PLoS Negl. Trop. Dis. 2014, 8, e3301. [Google Scholar] [CrossRef]
- Wang, X.; Liu, T.; Wu, Y.; Zhong, D.; Zhou, G.; Su, X.; Xu, J.; Sotero, C.F.; Sadruddin, A.A.; Wu, K.; et al. Bacterial Microbiota Assemblage in Aedes albopictus Mosquitoes and Its Impacts on Larval Development. Mol. Ecol. 2018, 27, 2972–2985. [Google Scholar] [CrossRef]
- Onyango, G.M.; Bialosuknia, M.S.; Payne, F.A.; Mathias, N.; Ciota, T.A.; Kramer, D.L. Increase in Temperature Enriches Heat Tolerant Taxa in Aedes aegypti Midguts. Sci. Rep. 2020, 10, 19135. [Google Scholar] [CrossRef]
- Onyango, M.G.; Lange, R.; Bialosuknia, S.; Payne, A.; Mathias, N.; Kuo, L.; Vigneron, A.; Nag, D.; Kramer, L.D.; Ciota, A.T. Zika Virus and Temperature Modulate Elizabethkingia Anophelis in Aedes albopictus. Parasites Vectors 2021, 14, 573. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.; Khanipov, K.; Albayrak, L.; Golovko, G.; Pimenova, M.; Saldaña, M.A.; Rojas, M.M.; Hornett, E.A.; Motl, G.C.; Fredregill, C.L.; et al. Microbiome Interaction Networks and Community Structure from Laboratory-Reared and Field-Collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus Mosquito Vectors. Front. Microbiol. 2018, 9, 2160. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, K.; Gunathilaka, N.; Amarasinghe, D.; Rodrigo, W.; Udayanga, L. Diversity of Midgut Bacteria in Larvae and Females of Aedes aegypti and Aedes albopictus from Gampaha District, Sri Lanka. Parasites Vectors 2021, 14, 433. [Google Scholar] [CrossRef] [PubMed]
- Tuanudom, R.; Yurayart, N.; Rodkhum, C.; Tiawsirisup, S. Diversity of Midgut Microbiota in Laboratory-Colonized and Field-Collected Aedes albopictus (Diptera: Culicidae): A Preliminary Study. Heliyon 2021, 7, e08259. [Google Scholar] [CrossRef]
- Conn, J.E.; Lainhart, W.; Rios, C.T.; Vinetz, J.M.; Bickersmith, S.A.; Moreno, M. Changes in Genetic Diversity from Field to Laboratory During Colonization of Anopheles darlingi Root (Diptera: Culicidae). Am. J. Trop. Med. Hyg. 2015, 93, 998–1001. [Google Scholar] [CrossRef]
- Muturi, E.J.; Njoroge, T.M.; Dunlap, C.; Cáceres, C.E. Blood Meal Source and Mixed Blood-Feeding Influence Gut Bacterial Community Composition in Aedes aegypti. Parasites Vectors 2021, 14, 83. [Google Scholar] [CrossRef]
- Sarma, D.K.; Kumar, M.; Dhurve, J.; Pal, N.; Sharma, P.; James, M.M.; Das, D.; Mishra, S.; Shubham, S.; Kumawat, M.; et al. Influence of Host Blood Meal Source on Gut Microbiota of Wild Caught Aedes aegypti, A Dominant Arboviral Disease Vector. Microorganisms 2022, 10, 332. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, D.; Augustinos, A.; Doudoumis, V.; Bel Mokhtar, N.; Maiga, H.; Tsiamis, G.; Bourtzis, K. Multiple Factors Determine the Structure of Bacterial Communities Associated with Aedes albopictus Under Artificial Rearing Conditions. Front. Microbiol. 2020, 11, 605. [Google Scholar] [CrossRef]
- Kain, M.P.; Skinner, E.B.; Athni, T.S.; Ramirez, A.L.; Mordecai, E.A.; van den Hurk, A.F. Not All Mosquitoes Are Created Equal: A Synthesis of Vector Competence Experiments Reinforces Virus Associations of Australian Mosquitoes. PLoS Negl. Trop. Dis. 2022, 16, e0010768. [Google Scholar] [CrossRef]
- Akorli, J.; Namaali, P.A.; Ametsi, G.W.; Egyirifa, R.K.; Pels, N.A.P. Generational Conservation of Composition and Diversity of Field-Acquired Midgut Microbiota in Anopheles gambiae (Sensu Lato) during Colonization in the Laboratory. Parasites Vectors 2019, 12, 27. [Google Scholar] [CrossRef]
- Boissière, A.; Tchioffo, M.T.; Bachar, D.; Abate, L.; Marie, A.; Nsango, S.E.; Shahbazkia, H.R.; Awono-Ambene, P.H.; Levashina, E.A.; Christen, R.; et al. Midgut Microbiota of the Malaria Mosquito Vector Anopheles gambiae and Interactions with Plasmodium Falciparum Infection. PLoS Pathog. 2012, 8, e1002742. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, E.V.; Hegde, S.; Roundy, C.M.; Golovko, G.; Saldaña, M.A.; Hart, C.E.; Anderson, E.R.; Hornett, E.A.; Khanipov, K.; Popov, V.L.; et al. Microbial Interactions in the Mosquito Gut Determine Serratia Colonization and Blood-Feeding Propensity. ISME J. 2021, 15, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Kersters, K.; Lisdiyanti, P.; Komagata, K.; Swings, J. The Family Acetobacteraceae: The Genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter, and Kozakia. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 163–200. [Google Scholar]
- Crotti, E.; Rizzi, A.; Chouaia, B.; Ricci, I.; Favia, G.; Alma, A.; Sacchi, L.; Bourtzis, K.; Mandrioli, M.; Cherif, A.; et al. Acetic Acid Bacteria, Newly Emerging Symbionts of Insects. Appl. Environ. Microbiol. 2010, 76, 6963–6970. [Google Scholar] [CrossRef] [PubMed]
- Saab, S.A.; Dohna, H.Z.; Nilsson, L.K.; Onorati, P.; Nakhleh, J.; Terenius, O.; Osta, M.A. The Environment and Species Affect Gut Bacteria Composition in Laboratory Co-Cultured Anopheles gambiae and Aedes albopictus Mosquitoes. Sci. Rep. 2020, 10, 3352. [Google Scholar] [CrossRef] [PubMed]
- Zug, R.; Hammerstein, P. Still a Host of Hosts for Wolbachia: Analysis of Recent Data Suggests That 40% of Terrestrial Arthropod Species Are Infected. PLoS ONE 2012, 7, 7–9. [Google Scholar] [CrossRef]
- Sinkins, S.P.; Braig, H.R.; Oneill, S.L. Wolbachia Pipientis: Bacterial Density and Unidirectional Cytoplasmic Incompatibility between Infected Populations of Aedes albopictus. Exp. Parasitol. 1995, 81, 284–291. [Google Scholar] [CrossRef]
- Zouache, K.; Voronin, D.; Tran-Van, V.; Mousson, L.; Failloux, A.B.; Mavingui, P. Persistent Wolbachia and Cultivable Bacteria Infection in the Reproductive and Somatic Tissues of the Mosquito Vector Aedes albopictus. PLoS ONE 2009, 4, e6388. [Google Scholar] [CrossRef]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M.; et al. Native Microbiome Impedes Vertical Transmission of Wolbachia in Anopheles Mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498–12503. [Google Scholar] [CrossRef]
- Rossi, P.; Ricci, I.; Cappelli, A.; Damiani, C.; Ulissi, U.; Mancini, M.V.; Valzano, M.; Capone, A.; Epis, S.; Crotti, E.; et al. Mutual Exclusion of Asaia and Wolbachia in the Reproductive Organs of Mosquito Vectors. Parasites Vectors 2015, 8, 278. [Google Scholar] [CrossRef]
- Tortosa, P.; Charlat, S.; Labbé, P.; Dehecq, J.S.; Barré, H.; Weill, M. Wolbachia Age-Sex-Specific Density in Aedes albopictus: A Host Evolutionary Response to Cytoplasmic Incompatibility? PLoS ONE 2010, 5, e9700. [Google Scholar] [CrossRef]
- Calvitti, M.; Marini, F.; Desiderio, A.; Puggioli, A.; Moretti, R. Wolbachia Density and Cytoplasmic Incompatibility in Aedes albopictus: Concerns with Using Artificial Wolbachia Infection as a Vector Suppression Tool. PLoS ONE 2015, 10, e0121813. [Google Scholar] [CrossRef] [PubMed]
- Mancini, M.V.; Herd, C.S.; Ant, T.H.; Murdochy, S.M.; Sinkins, S.P. Wolbachia Strain WAu Efficiently Blocks Arbovirus Transmission in Aedes albopictus. PLoS Negl. Trop. Dis. 2020, 14, e0007926. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.H.M.; Gonçalves, R.L.S.; Lara, F.A.; Dias, F.A.; Gandara, A.C.P.; Menna-Barreto, R.F.S.; Edwards, M.C.; Laurindo, F.R.M.; Silva-Neto, M.A.C.; Sorgine, M.H.F.; et al. Blood Meal-Derived Heme Decreases ROS Levels in the Midgut of Aedes aegypti and Allows Proliferation of Intestinal Microbiota. PLoS Pathog. 2011, 7, e1001320. [Google Scholar] [CrossRef] [PubMed]
- Valiente Moro, C.; Tran, F.H.; Nantenaina Raharimalala, F.; Ravelonandro, P.; Mavingui, P. Diversity of Culturable Bacteria Including Pantoea in Wild Mosquito Aedes albopictus. BMC Microbiol. 2013, 13, 70. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Lu, C.-W.; Shyu, Y.-T.; Lin, S.-S. Revealing the Saline Adaptation Strategies of the Halophilic Bacterium Halomonas Beimenensis through High-Throughput Omics and Transposon Mutagenesis Approaches. Sci. Rep. 2017, 7, 13037. [Google Scholar] [CrossRef]
- Rani, A.; Sharma, A.; Rajagopal, R.; Adak, T.; Bhatnagar, R.K. Bacterial Diversity Analysis of Larvae and Adult Midgut Microflora Using Culture-Dependent and Culture-Independent Methods in Lab-Reared and Field-Collected Anopheles stephensi-an Asian Malarial Vector. BMC Microbiol. 2009, 9, 96. [Google Scholar] [CrossRef]
- David, M.R.; Maria, L.; Carolina, A.; Vicente, P.; Maciel-de-freitas, R. Effects of Environment, Dietary Regime and Ageing on the Dengue Vector Microbiota: Evidence of a Core Microbiota throughout Aedes aegypti Lifespan. Memórias Do Inst. Oswaldo Cruz 2016, 111, 577–587. [Google Scholar] [CrossRef]
- Rosso, F.; Tagliapietra, V.; Albanese, D.; Pindo, M.; Baldacchino, F.; Arnoldi, D.; Donati, C.; Rizzoli, A. Reduced Diversity of Gut Microbiota in Two Aedes Mosquitoes Species in Areas of Recent Invasion. Sci. Rep. 2018, 8, 16091. [Google Scholar] [CrossRef]
- Chavshin, A.R.; Oshaghi, M.A.; Vatandoost, H.; Yakhchali, B.; Zarenejad, F. Malpighian Tubules Are Important Determinants of Pseudomonas Transstadial Transmission and Longtime Persistence in Anopheles stephensi. Parasites Vectors 2015, 8, 585. [Google Scholar] [CrossRef]
- Minard, G.; Tran, F.H.; Raharimalala, F.N.; Hellard, E.; Ravelonandro, P.; Mavingui, P.; Valiente Moro, C. Prevalence, Genomic and Metabolic Profiles of Acinetobacter and Asaia Associated with Field-Caught Aedes albopictus from Madagascar. FEMS Microbiol. Ecol. 2013, 83, 63–73. [Google Scholar] [CrossRef]
- Colquhoun, J.A.; Heald, S.C.; Li, L.; Tamaoka, J.; Kato, C.; Horikoshi, K.; Bull, A.T. Taxonomy and Biotransformation Activities of Some Deep-Sea Actinomycetes. Extremophiles 1998, 2, 269–277. [Google Scholar] [CrossRef] [PubMed]
- George, M.; Anjumol, A.; George, G.; Mohamed Hata, A.A. Distribution and Bioactive Potential of Soil Actinomycetes from Different Ecological Habitats. Afr. J. Microbiol. Res. 2012, 6, 2265–2271. [Google Scholar] [CrossRef]
- Chaudhary, H.S.; Soni, B.; Shrivastava, A.R.; Shrivastava, S. Diversity and Versatility of Actinomycetes and Its Role in Antibiotic Production. J. Appl. Pharm. Sci. 2013, 3, S83–S94. [Google Scholar] [CrossRef]
- Dada, N.; Jumas-Bilak, E.; Manguin, S.; Seidu, R.; Stenström, T.A.; Overgaard, H.J. Comparative Assessment of the Bacterial Communities Associated with Aedes aegypti Larvae and Water from Domestic Water Storage Containers. Parasites Vectors 2014, 7, 391. [Google Scholar] [CrossRef]
- Dahmana, H.; Sambou, M.; Raoult, D.; Fenollar, F.; Mediannikov, O. Biological Control of Aedes albopictus: Obtained from the New Bacterial Candidates with Insecticidal Activity. Insects 2020, 11, 403. [Google Scholar] [CrossRef]
- Lindh, J.M.; Kännaste, A.; Knols, B.G.J.; Faye, I.; Borg-Karlson, A.K. Oviposition Responses of Anopheles gambiae s.s. (Diptera: Culicidae) and Identification of Volatiles from Bacteria-Containing Solutions. J. Med. Entomol. 2008, 45, 1039–1049. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Diversity and Antibiotic Resistance Patterns of Sphingomonadaceae Isolates from Drinking Water. Appl. Environ. Microbiol. 2011, 77, 5697–5706. [Google Scholar] [CrossRef]
- Mandic-Mulec, I.; Stefanic, P.; van Elsas, J.D. Ecology of Bacillaceae. Microbiol. Spectr. 2015, 3, 59–85. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Van Wyk, J.; Morkel, R.A.; Dolley, L. Metabolites of Propionibacterium: Techno- and Biofunctional Ingredients. Altern. Replace. Foods 2018, 17, 205–260. [Google Scholar] [CrossRef]

| Sample Classification | Collection Area | Collection Season | Origin | Generation |
|---|---|---|---|---|
| JDG-W | Jardim Guanabara | Wet | Field | F0 |
| JU-W | Jurujuba | Wet | Field | F0 |
| RPC-W | Represa dos Ciganos | Wet | Field | F0 |
| JDG-D | Jardim Guanabara | Dry | Field | F0 |
| JU-D | Jurujuba | Dry | Field | F0 |
| RPC-D | Represa dos Ciganos | Dry | Field | F0 |
| JDG-F1 | Jardim Guanabara | LAB * | Laboratory | F1 |
| JU-F1 | Jurujuba | LAB * | Laboratory | F1 |
| RPC-F1 | Represa dos Ciganos | LAB * | Laboratory | F1 |
| LAB | Laboratory | LAB * | Laboratory | >F30 |
| Variable | Description | Groups (In Bold) |
|---|---|---|
| Area | Samples classified according to collection area (regardless of the collection season) and its respective F1. | JDG (JDG-W + JDG-D), JU (JU-W + JU-D), RPC (RPC-W+RPC-D), JDG-F1, JU-F1, RPC-F1 vs. LAB |
| Collection season | Period in which insects were collected (regardless of collection area) | WET (JDG-W + JU-W + RPC-W) vs. DRY (JDG-D + JU-D + RPC-D); F1 (JDG-F1 + JU-F1 + RPC-F1) vs. LAB |
| Origin | Place of mosquito rearing (field or laboratory, regardless collection area and season) | FIELD (JDG-W + JU-W + RPC-W + JDG-D + JU-D + RPC-D), F1 (JDG-F1 + JU-F1 + RPC-F1) vs. LAB |
| Population | Mosquito population (regardless of collection area, season or rearing place) | JDG (JDG-W + JDG-D + JDG-F1), JU (JU-W + JU-D + JU-F1), RPC (RPC-W + RPC-D + RPC-F1) vs. LAB |
| Significantly Different Taxa | W * | F1 | FIELD | LAB |
|---|---|---|---|---|
| Propionibacterium | 202 |  |  | |
| Acetobacteraceae_unclassified genus | 202 |  |  | |
| Alphaproteobacteria_unclassified order | 200 |  |  | |
| Peptostreptococcaceae_unclassified genus | 196 |  |  | |
| Rahnella | 188 |  |  | |
| Cupriavidus | 187 |  | ||
| Moraxellaceae_unclassified genus | 186 |  | ||
| Pedobacter | 185 |  | ||
| Reyranella | 182 |  | ||
| MLE1-12_unclassified family | 182 |  | ||
| Rhodospirillaceae_unclassified genus | 176 |  |  | |
| Hydrocarboniphaga | 175 |  | ||
| Sphingobacteriales_unclassified family | 172 |  |  |
| Significantly Different Taxa | W * | WET | DRY | F1 | LAB |
|---|---|---|---|---|---|
| Propionibacterium | 202 |  |  |  | |
| Acetobacteraceae_unclassified genus | 202 |  |  |  | |
| Wolbachia | 202 |  |  |  | |
| Methylobacterium | 202 |  | |||
| Peptostreptococcaceae_unclassified genus | 201 |  |  |  | |
| Alphaproteobacteria_unclassified order | 199 |  |  |  | |
| Methylobacteriaceae_unclassified genus | 193 |  |  | ||
| Cupriavidus | 190 |  | |||
| Moraxellaceae_unclassified genus | 187 |  | |||
| Pedobacter | 183 |  | |||
| Reyranella | 183 |  | |||
| MLE1-12_unclassified family | 183 |  | |||
| Rahnella | 177 |  |  | ||
| Rhizobiales_unclassified family | 177 |  |  |  | |
| Hydrocarboniphaga | 177 |  | |||
| Rhodospirillaceae_unclassified genus | 174 |  |  | ||
| Sphingomonadaceae_unclassified genus | 172 |  |  |  | |
| Sphingobacteriales_unclassified family | 171 |  |  | ||
| Bdellovibrio | 171 |  | |||
| Ralstonia | 171 |  |  |  | |
| Chitinophagaceae_unclassified genus | 169 |  |
| Population | IR—14dpi (%) | IR—21dpi (%) | DR—14dpi (%) | DR—21dpi (%) |
|---|---|---|---|---|
| JDG-F1 | 62.5 (25/40) | 40 (4/10) | 16 (4/25) | 25 (1/4) |
| JU-F1 | 30 (12/40) | 44.4 (24/54) | 16.7 (2/12) | 45.8 (11/24) |
| RPC-F1 | 85 (34/40) | 85.4 (35/41) | 64.7 (22/34) | 71.4 (25/35) |
| Ae. aegypti (control) | 100 (20/20) | 95 (19/20) | 95 (19/20) | 100 (19/19) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baltar, J.M.C.; Pavan, M.G.; Corrêa-Antônio, J.; Couto-Lima, D.; Maciel-de-Freitas, R.; David, M.R. Gut Bacterial Diversity of Field and Laboratory-Reared Aedes albopictus Populations of Rio de Janeiro, Brazil. Viruses 2023, 15, 1309. https://doi.org/10.3390/v15061309
Baltar JMC, Pavan MG, Corrêa-Antônio J, Couto-Lima D, Maciel-de-Freitas R, David MR. Gut Bacterial Diversity of Field and Laboratory-Reared Aedes albopictus Populations of Rio de Janeiro, Brazil. Viruses. 2023; 15(6):1309. https://doi.org/10.3390/v15061309
Chicago/Turabian StyleBaltar, João M. C., Márcio G. Pavan, Jessica Corrêa-Antônio, Dinair Couto-Lima, Rafael Maciel-de-Freitas, and Mariana R. David. 2023. "Gut Bacterial Diversity of Field and Laboratory-Reared Aedes albopictus Populations of Rio de Janeiro, Brazil" Viruses 15, no. 6: 1309. https://doi.org/10.3390/v15061309
APA StyleBaltar, J. M. C., Pavan, M. G., Corrêa-Antônio, J., Couto-Lima, D., Maciel-de-Freitas, R., & David, M. R. (2023). Gut Bacterial Diversity of Field and Laboratory-Reared Aedes albopictus Populations of Rio de Janeiro, Brazil. Viruses, 15(6), 1309. https://doi.org/10.3390/v15061309








