Proteomics Identified UDP-Glycosyltransferase Family Members as Pro-Viral Factors for Turnip Mosaic Virus Infection in Nicotiana benthamiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Virus Inoculation
2.2. Protein Extraction, Trypsin Digestion, TMT/iTRAQ Labelling and LC-MS/MS Analysis
2.3. Database Search and Bioinformatic Analysis
2.4. RNA Extraction and RT-qPCR
2.5. Gene Cloning and Plasmid Construction
2.6. Western Blotting
3. Results
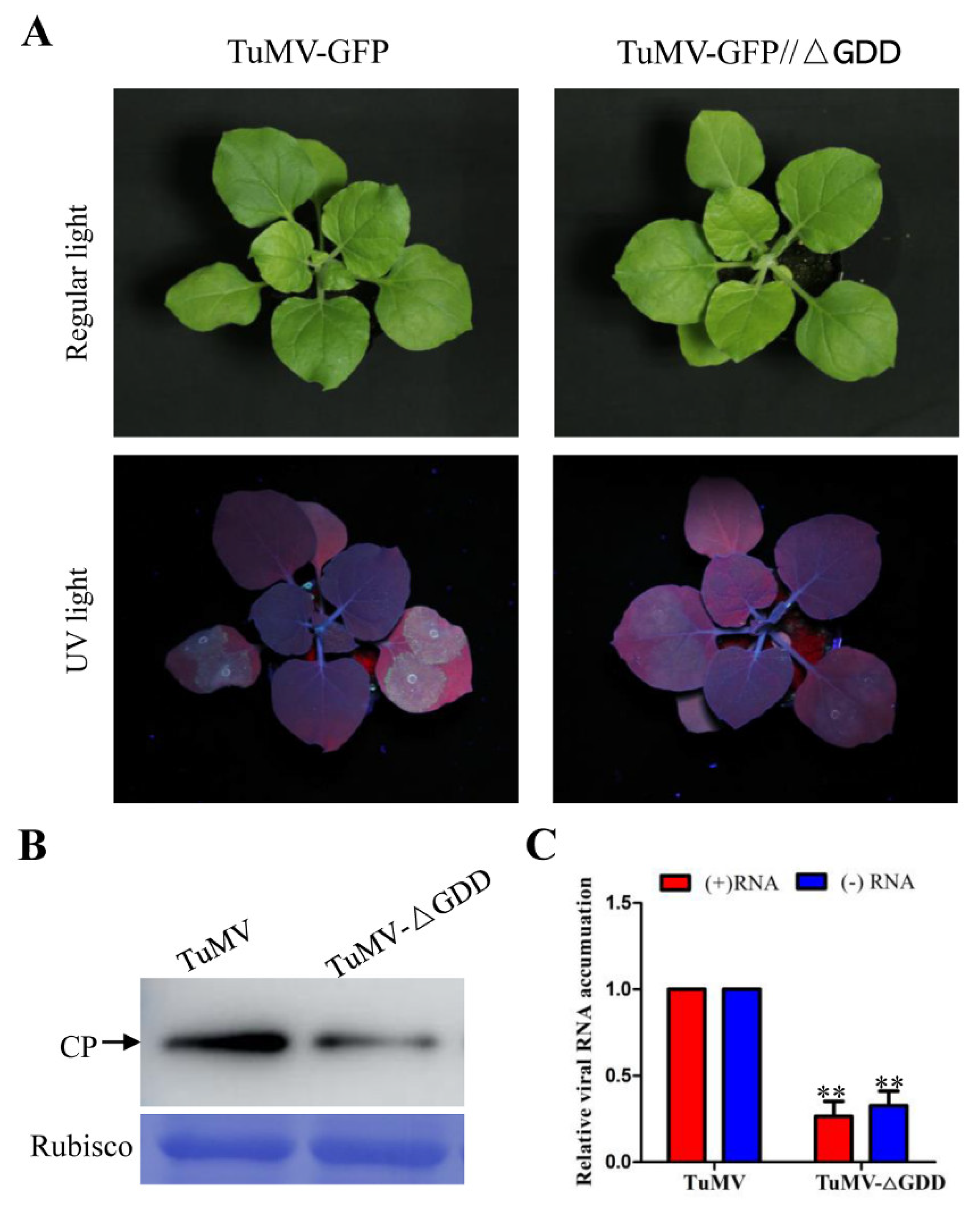
3.1. Wild Type and Replication-Deficient TuMV Infection
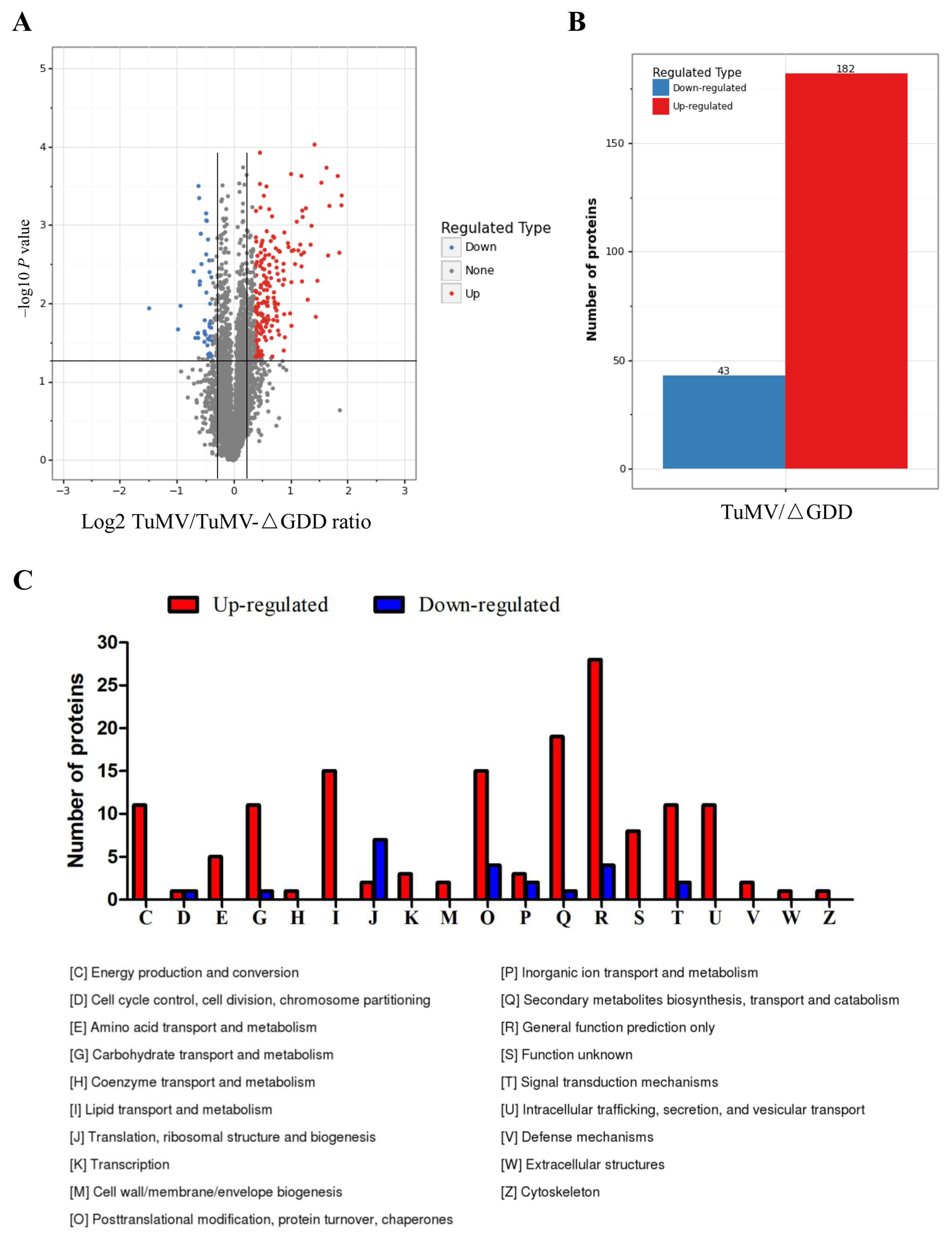
3.2. Identification and Quantification of Total Proteins by iTRAQ
3.3. GO Enrichment Analyses of Host DAPs
3.4. Validation of UDP- Glycosyltransferases (UGTs) Proteins Using RT-qPCR
3.5. Virus-Induced Gene Silencing Screen Revealed That NbUGT91C1 and NbUGT74F1 Are Required for TuMV Infection
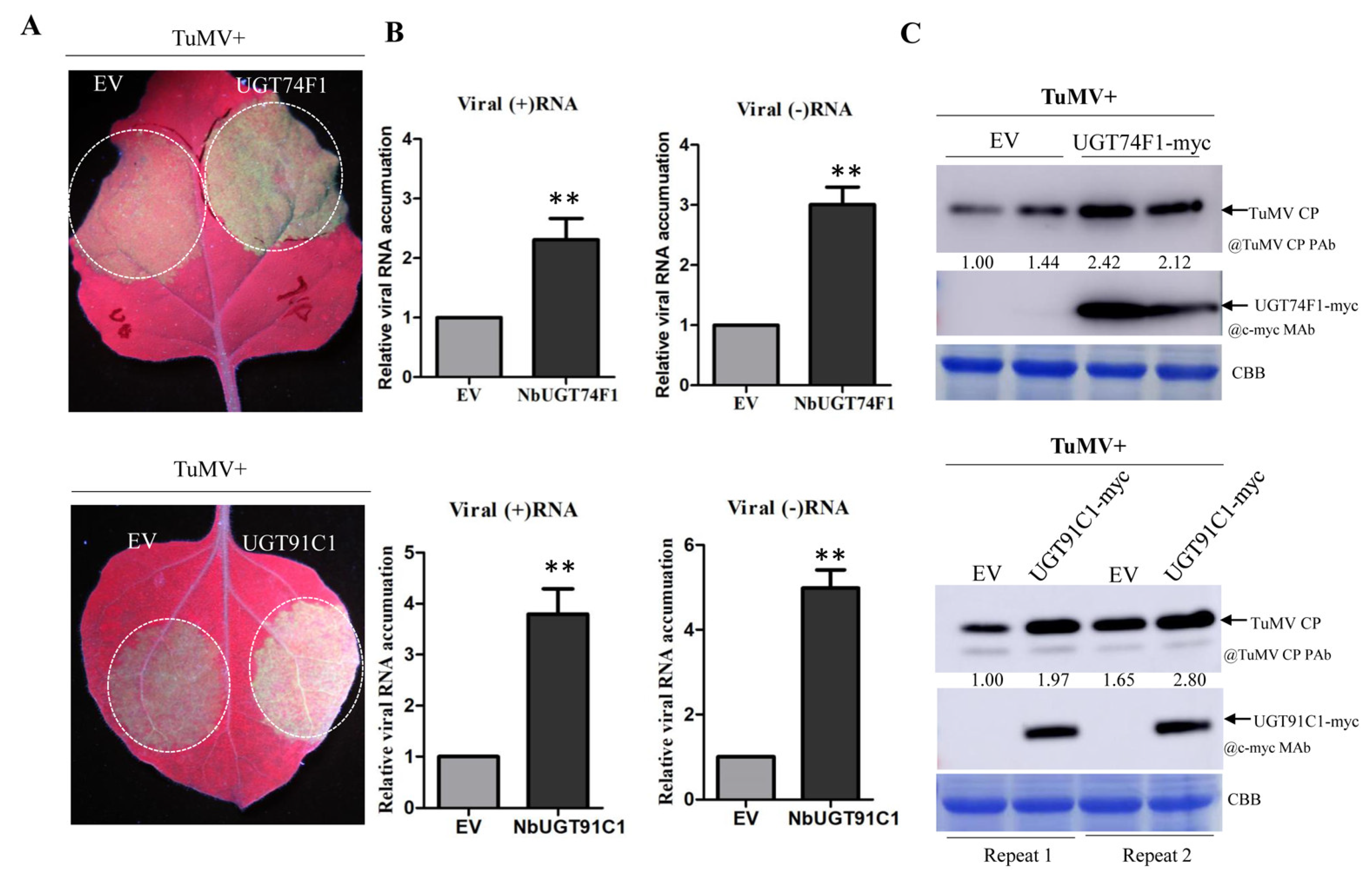
3.6. Over-Expression of NbUGT74F1 or NbUGT91C1 Promotes TuMV Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vélez-Bermúdez, I.C.; Wen, T.-N.; Lan, P.; Schmidt, W. Isobaric tag for relative and absolute Quantitation (iTRAQ)-Based Protein Profiling in Plants. In Plant Proteostasis: Methods and Protocols; Lois, L.M., Matthiesen, R., Eds.; Springer: New York, NY, USA, 2016; pp. 213–221. [Google Scholar]
- Bowles, D.; Isayenkova, J.; Lim, E.K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Lim, E.-K. Glycosyltransferases of small molecules: Their roles in plant biology. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010. [Google Scholar] [CrossRef]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Mateo-Bonmati, E.; Casanova-Saez, R.; Simura, J.; Ljung, K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New. Phytol. 2021, 232, 642–654. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, M.; Lu, M.; Wu, Y.; Jing, T.; Zhao, M.; Zhao, Y.; Feng, Y.; Wang, J.; Gao, T.; et al. Salicylic acid carboxyl glucosyltransferase UGT87E7 regulates disease resistance in Camellia sinensis. Plant Physiol. 2022, 188, 1507–1520. [Google Scholar] [CrossRef]
- He, Y.; Wu, L.; Liu, X.; Jiang, P.; Yu, L.; Qiu, J.; Wang, G.; Zhang, X.; Ma, H. TaUGT6, a novel UDP-Glycosyltransferase gene enhances the resistance to FHB and DON accumulation in wheat. Front. Plant Sci. 2020, 11, 574775. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Q.; Liu, X.; Cheng, J.; Zhang, L.; Chu, H.; Wang, R.; Li, H.; Chang, H.; Ahmed, N.; et al. pUGTdb: A comprehensive database of plant UDP-dependent glycosyltransferases. Mol. Plant 2023, 16, 643–646. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, W.; Shan, X.; Liu, J.; Zhao, L.; Zhao, Q. Glycoside-specific metabolomics combined with precursor isotopic labeling for characterizing plant glycosyltransferases. Mol. Plant 2022, 15, 1517–1532. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Wang, A. Research advances in potyviruses: From the laboratory bench to the field. Annu. Rev. Phytopathol. 2021, 59, 1–29. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Jordan, R.; Kreuze, J.; Li, F.; López-Moya, J.J.; Mäkinen, K.; Ohshima, K.; Wylie, S.J.; ICTV Report Consortium. ICTV virus taxonomy profile: Potyviridae 2022. J. Gen. Virol. 2022, 103, 001738. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; Garcia, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar] [PubMed]
- Rodamilans, B.; Valli, A.; Garcia, J.A. Molecular plant-plum pox virus interactions. Mol. Plant Microbe Interact. 2020, 33, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.; Carrasco, J.L.; Toft, C.; Hillung, J.; Gimenez-Santamarina, S.; Yenush, L.; Rodrigo, G.; Elena, S.F. A binary interaction map between turnip mosaic virus and Arabidopsis thaliana proteomes. Commun. Biol. 2023, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Nellist, C.F.; Ohshima, K.; Ponz, F.; Walsh, J.A. Turnip mosaic virus, a virus for all seasons. Ann. Appl. Biol. 2022, 180, 312–327. [Google Scholar] [CrossRef]
- Wu, G.W.; Cui, X.Y.; Chen, H.; Renaud, J.B.; Yu, K.F.; Chen, X.; Wang, A.M. Dynamin-like proteins of endocytosis in plants are coopted by Potyviruses to enhance virus Infection. J. Virol. 2018, 92, e01320-18. [Google Scholar] [CrossRef]
- Wu, G.W.; Jia, Z.X.; Ding, K.D.; Zheng, H.Y.; Lu, Y.W.; Lin, L.; Peng, J.J.; Rao, S.F.; Wang, A.M.; Chen, J.P.; et al. Turnip mosaic virus co-opts the vacuolar sorting receptor VSR4 to promote viral genome replication in plants by targeting viral replication vesicles to the endosome. PLoS Pathog. 2022, 18, e1010257. [Google Scholar] [CrossRef]
- Cheng, X.; Xiong, R.; Li, Y.; Li, F.; Zhou, X.; Wang, A. Sumoylation of turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 2017, 29, 508–525. [Google Scholar] [CrossRef]
- Ji, M.; Zhao, J.; Han, K.; Cui, W.; Wu, X.; Chen, B.; Lu, Y.; Peng, J.; Zheng, H.; Rao, S.; et al. Turnip mosaic virus P1 suppresses JA biosynthesis by degrading cpSRP54 that delivers AOCs onto the thylakoid membrane to facilitate viral infection. PLoS Pathog. 2021, 17, e1010108. [Google Scholar] [CrossRef]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberte, J.F. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef]
- Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-dependent RNA polymerase NIb of potyviruses plays multifunctional, contrasting roles during viral infection. Viruses 2020, 12, 77. [Google Scholar] [CrossRef]
- Cui, X.Y.; Yaghmaiean, H.; Wu, G.W.; Wu, X.Y.; Chen, X.; Thorn, G.; Wang, A.M. The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex. Virology 2017, 510, 147–155. [Google Scholar] [CrossRef]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Rosli, H.G.; Martin, G.B.; Mueller, L.A. The SGN VIGS tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant 2015, 8, 486–488. [Google Scholar] [CrossRef]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Lum, K.K.; Cristea, I.M. Proteomic approaches to uncovering virus–host protein interactions during the progression of viral infection. Expert Rev. Proteom. 2016, 13, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Das, P.P.; Oppenheimer, P.; Zhou, G.; Wong, S.M. iTRAQ-based protein analysis provides insight into heterologous superinfection exclusion with TMV-43A against CMV in tobacco (Nicotiana benthamiana) plants. J. Proteomics 2020, 229, 103948. [Google Scholar] [CrossRef]
- Macharia, M.; Das, P.P.; Naqvi, N.I.; Wong, S.-M. iTRAQ-based quantitative proteomics reveals a ferroptosis-like programmed cell death in plants infected by a highly virulent tobacco mosaic virus mutant 24A+UPD. Phytopathol. Res. 2020, 2, 1. [Google Scholar] [CrossRef]
- Liu, H.; Das, P.P.; Zhang, J.; Yu, L.; Wang, M.; Lin, Q.; Zhou, Y.; Xu, Q.; Wong, S.M. iTRAQ-based quantitative proteomics suggests mitophagy involvement after Rice black-streaked dwarf virus acquisition in insect vector small brown planthopper Laodelphax striatellus Fallen. J. Proteomics 2021, 246, 104314. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.S.; Kwon, C. Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 2017, 40, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.W.; Cui, X.Y.; Dai, Z.J.; He, R.R.; Li, Y.Z.; Yu, K.F.; Bernards, M.; Chen, X.; Wang, A.M. A plant RNA virus hijacks endocytic proteins to establish its infection in plants. Plant J. 2020, 101, 384–400. [Google Scholar] [CrossRef]
- Wu, G.W.; Jia, Z.X.; Rui, P.H.; Zheng, H.Y.; Lu, Y.W.; Lin, L.; Peng, J.J.; Rao, S.F.; Wang, A.M.; Chen, J.P.; et al. Acidic dileucine motifs in the cylindrical inclusion protein of turnip mosaic virus are crucial for endosomal targeting and viral replication. Mol. Plant Pathol. 2022, 23, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 3, 342–352. [Google Scholar] [CrossRef]
- Restrepo-Hartwig, M.A.; Carrington, J.C. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 1994, 68, 2388–2397. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberte, J.F.; Zheng, H. Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef]
- Moller, I.M.; Sweetlove, L.J. ROS signalling—Specificity is required. Trends Plant Sci. 2010, 15, 370–374. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic acid: Biosynthesis and signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Ding, Y. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.X.; Zhu, G.Q.; Liu, Q.; Chen, L.; Li, Y.J.; Hou, B.K. Modulation of plant salicylic acid-associated immune responses via glycosylation of dihydroxybenzoic acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Noutoshi, Y.; Okazaki, M.; Kida, T.; Nishina, Y.; Morishita, Y.; Ogawa, T.; Suzuki, H.; Shibata, D.; Jikumaru, Y.; Hanada, A.; et al. Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 2012, 24, 3795–3804. [Google Scholar] [CrossRef]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant 2008, 132, 417–425. [Google Scholar] [CrossRef] [PubMed]
- George Thompson, A.M.; Iancu, C.V.; Neet, K.E.; Dean, J.V.; Choe, J.Y. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci. Rep. 2017, 7, 46629. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar] [CrossRef]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, K.; Jia, Z.; Rui, P.; Fang, X.; Zheng, H.; Chen, J.; Yan, F.; Wu, G. Proteomics Identified UDP-Glycosyltransferase Family Members as Pro-Viral Factors for Turnip Mosaic Virus Infection in Nicotiana benthamiana. Viruses 2023, 15, 1401. https://doi.org/10.3390/v15061401
Ding K, Jia Z, Rui P, Fang X, Zheng H, Chen J, Yan F, Wu G. Proteomics Identified UDP-Glycosyltransferase Family Members as Pro-Viral Factors for Turnip Mosaic Virus Infection in Nicotiana benthamiana. Viruses. 2023; 15(6):1401. https://doi.org/10.3390/v15061401
Chicago/Turabian StyleDing, Kaida, Zhaoxing Jia, Penghuan Rui, Xinxin Fang, Hongying Zheng, Jianping Chen, Fei Yan, and Guanwei Wu. 2023. "Proteomics Identified UDP-Glycosyltransferase Family Members as Pro-Viral Factors for Turnip Mosaic Virus Infection in Nicotiana benthamiana" Viruses 15, no. 6: 1401. https://doi.org/10.3390/v15061401
APA StyleDing, K., Jia, Z., Rui, P., Fang, X., Zheng, H., Chen, J., Yan, F., & Wu, G. (2023). Proteomics Identified UDP-Glycosyltransferase Family Members as Pro-Viral Factors for Turnip Mosaic Virus Infection in Nicotiana benthamiana. Viruses, 15(6), 1401. https://doi.org/10.3390/v15061401





