Dengue Virus Infection of Human Retinal Müller Glial Cells
Abstract
1. Introduction
2. Methods
2.1. Human Retinal Cell Lines
2.2. Primary Retinal Human Müller Cells
2.3. Dengue Virus Strains
2.4. Dengue Virus Infection of Human Retinal Müller Cells
2.5. Cell Immunolabelling
2.6. RNA Extraction and cDNA Synthesis
2.7. Quantitative Real-Time Polymerase Chain Reaction
2.8. Interferon-Beta Immunoassay
2.9. Statistical Analyses
3. Results
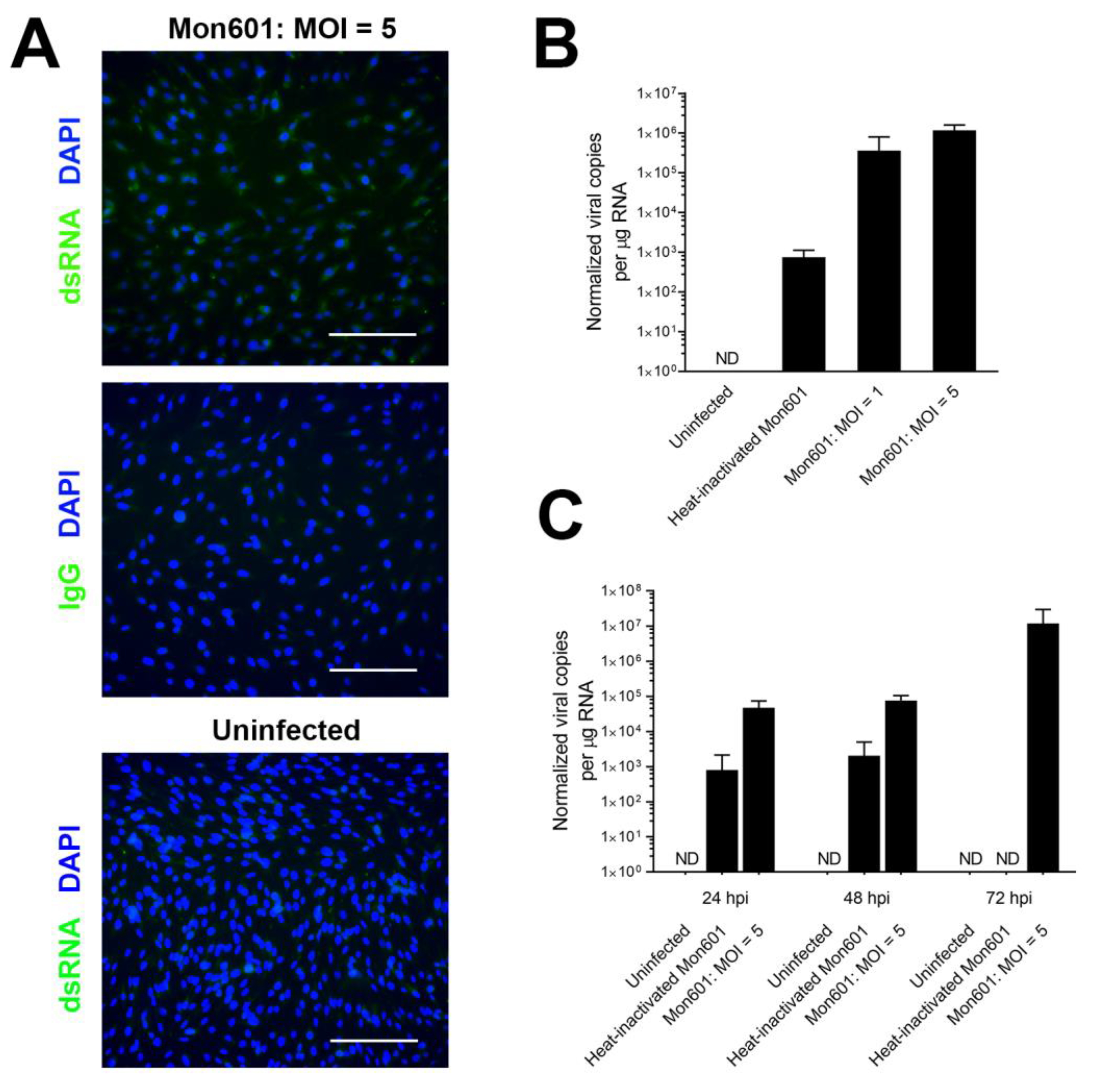
3.1. Susceptibility of the MIO-M1 Human Müller Cell Line to Infection with Dengue Virus
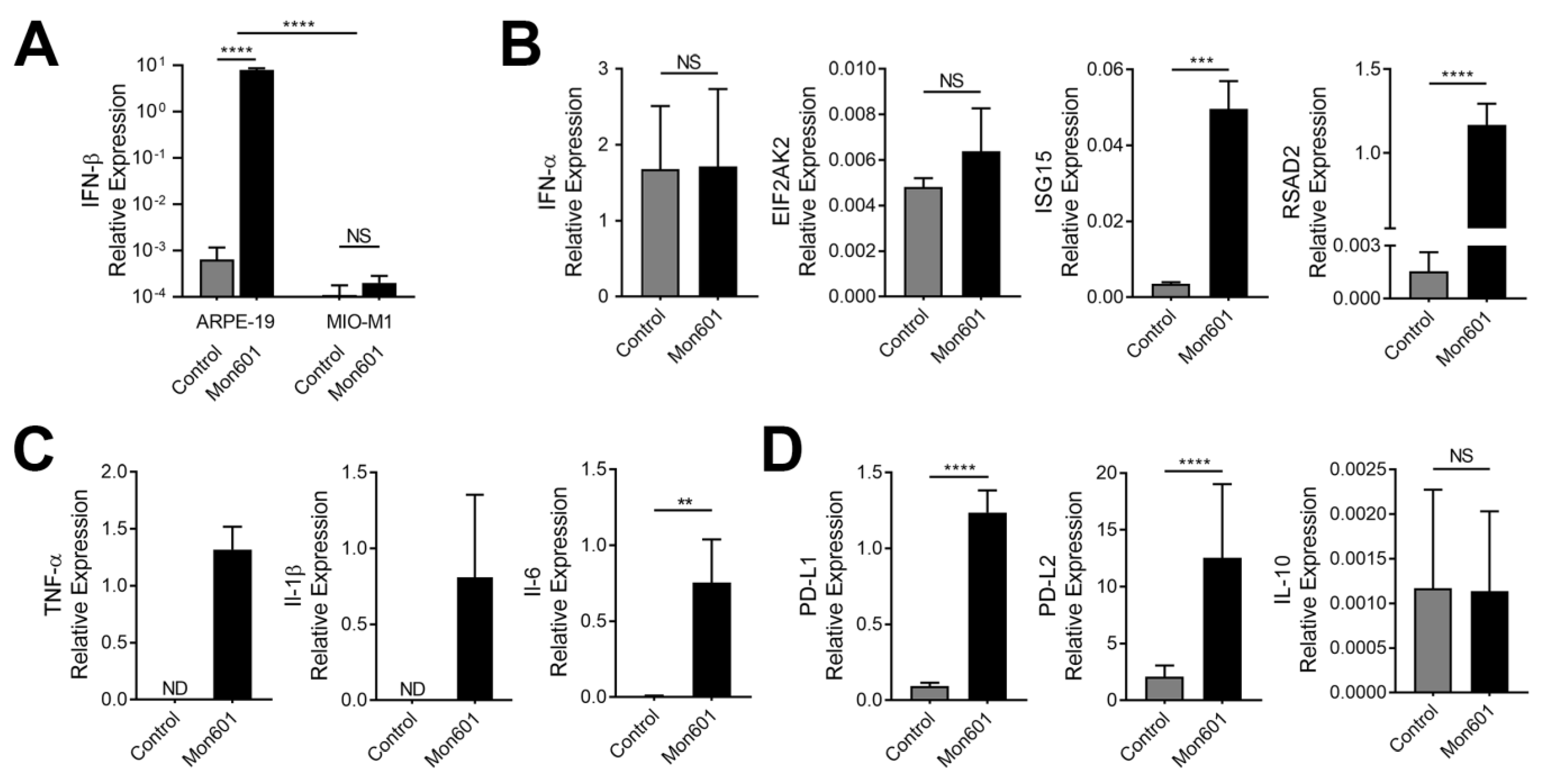
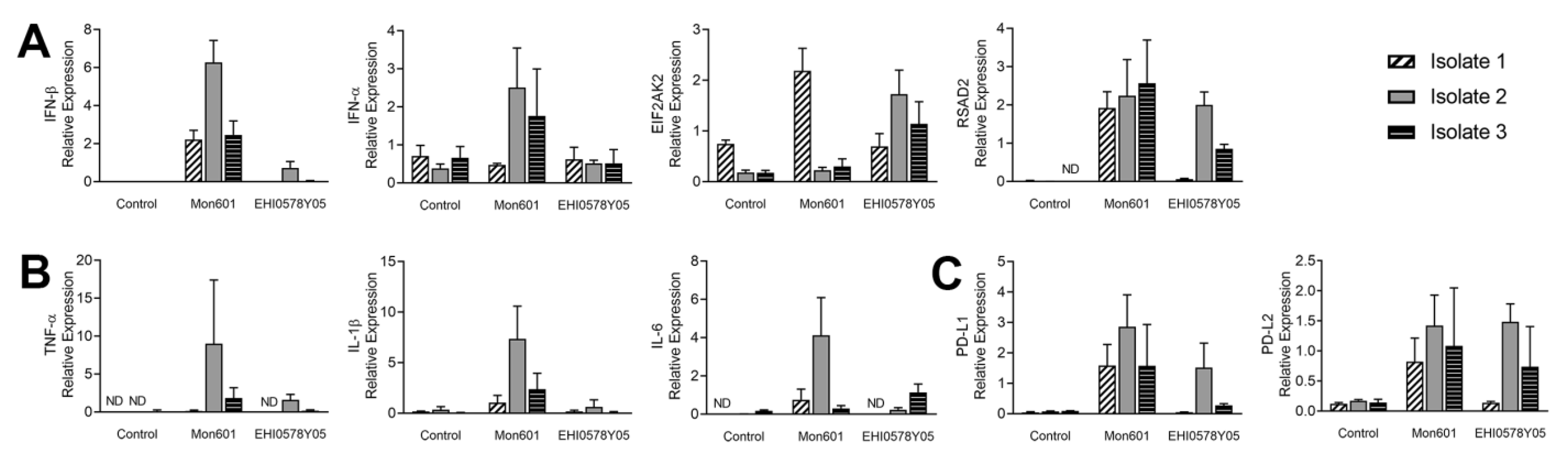
3.2. Type I Interferon Response in Dengue Virus-Infected MIO-M1 Human Müller Cells
3.3. Immune Responses of MIO-M1 Human Müller Cells to Infection with Dengue Virus
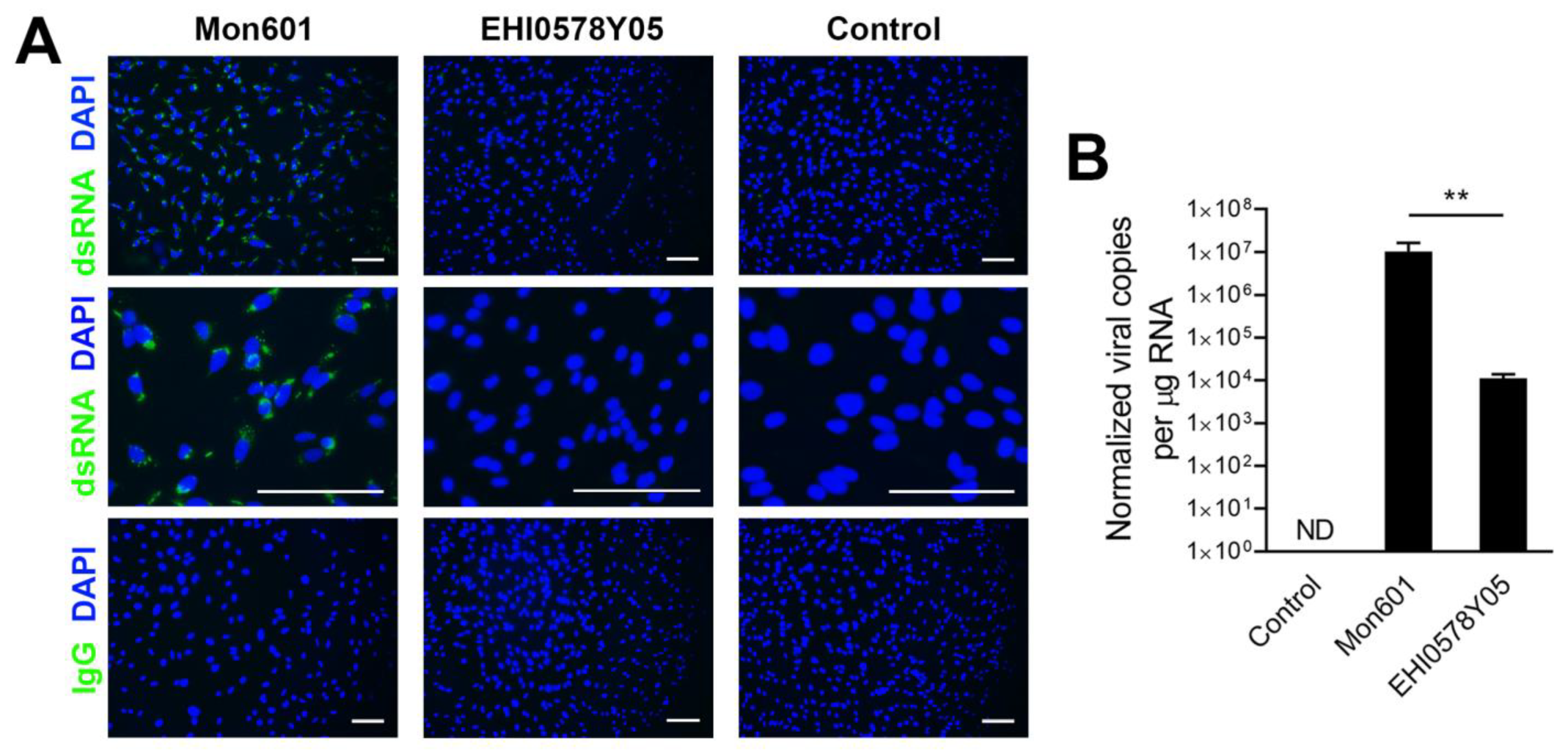
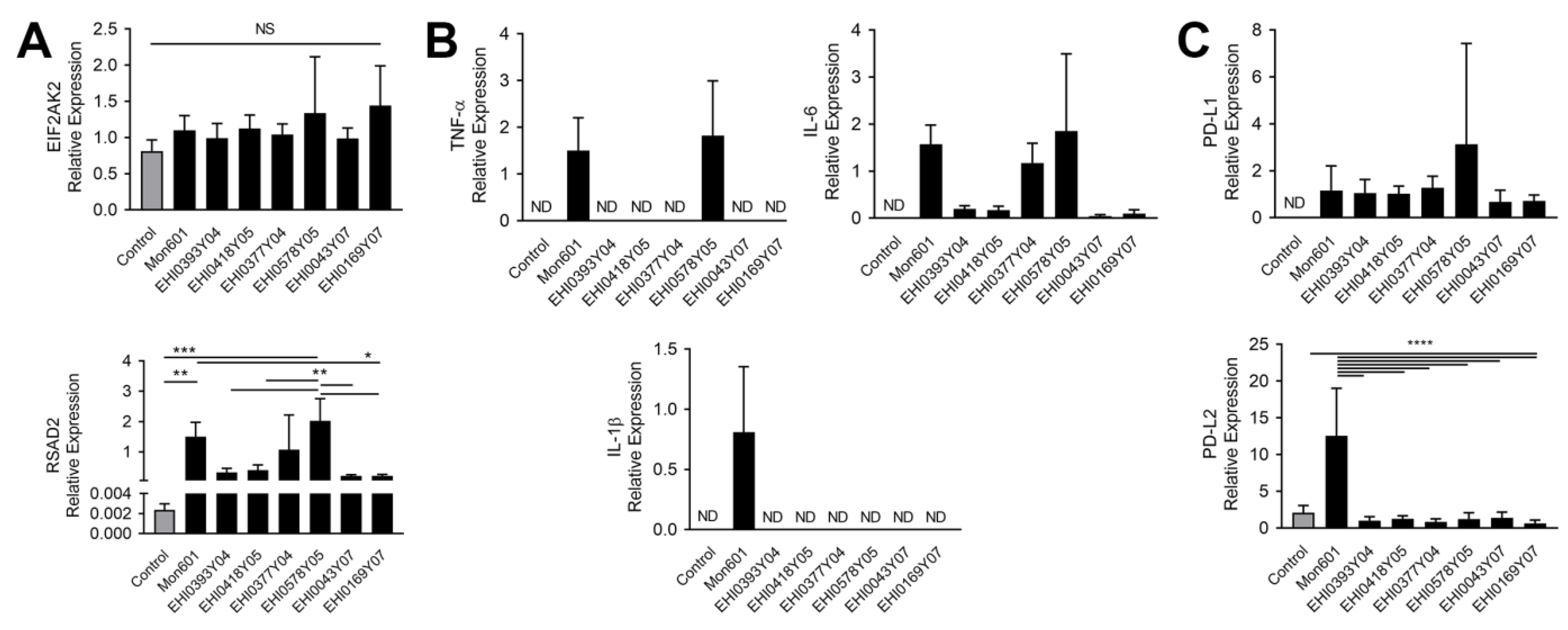
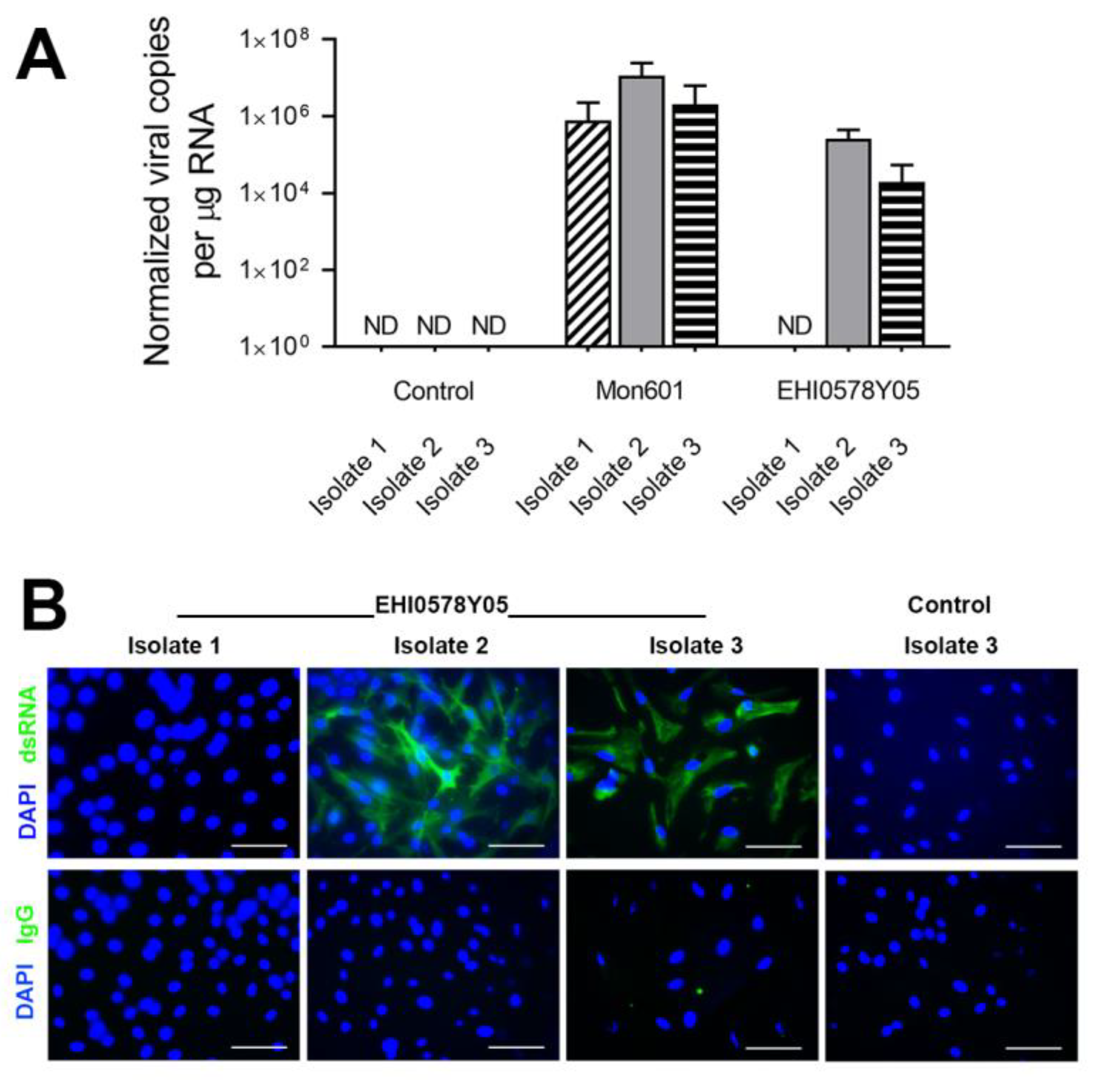
3.4. Infection of MIO-M1 Human Müller Cells with Dengue Virus Field Isolates
3.5. Responses to Dengue Virus Infection in Primary Human Müller Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roy, R.N. Affections of the eye following dengue. Ind. Med. Gaz. 1873, 8, 42–43. [Google Scholar] [PubMed]
- Gill, W. Ocular symptoms in dengue based on analysis of 1241 cases. Arch. Ophthalmol. 1928, 57, 628–638. [Google Scholar]
- Weymann, M.F. Some cases of ocular complication during an epidemic of dengue fever. Am. J. Ophthalmol. 1929, 12, 708. [Google Scholar]
- Weymann, M.F. The ocular complications of dengue fever during the last epidemic in Greece. Am. J. Ophthalmol. 1929, 12, 708. [Google Scholar]
- Richardson, S. Ocular symptoms and complications observed in dengue. Trans. Am. Ophthalmol. Soc. 1933, 31, 450–477. [Google Scholar]
- Chan, D.P.; Teoh, S.C.; Tan, C.S.; Nah, G.K.; Rajagopalan, R.; Prabhakaragupta, M.K.; Chee, C.K.; Lim, T.H.; Goh, K.Y. Ophthalmic complications of dengue. Emerg. Infect. Dis. 2006, 12, 285–289. [Google Scholar] [CrossRef]
- Kapoor, H.K.; Bhai, S.; John, M.; Xavier, J. Ocular manifestations of dengue fever in an East Indian epidemic. Can. J. Ophthalmol. 2006, 41, 741–746. [Google Scholar] [CrossRef]
- Su, D.H.; Bacsal, K.; Chee, S.P.; Flores, J.V.; Lim, W.K.; Cheng, B.C.; Jap, A.H.; Dengue Maculopathy Study Group. Prevalence of dengue maculopathy in patients hospitalized for dengue fever. Ophthalmology 2007, 114, 1743–1747. [Google Scholar] [CrossRef]
- Oliver, G.F.; Carr, J.M.; Smith, J.R. Emerging infectious uveitis: Chikungunya, dengue, Zika and Ebola: A review. Clin. Exp. Ophthalmol. 2019, 47, 372–380. [Google Scholar] [CrossRef]
- Chia, A.; Luu, C.D.; Mathur, R.; Cheng, B.; Chee, S.P. Electrophysiological findings in patients with dengue-related maculopathy. Arch. Ophthalmol. 2006, 124, 1421–1426. [Google Scholar] [CrossRef]
- Haydinger, C.D.; Ferreira, L.B.; Williams, K.A.; Smith, J.R. Mechanisms of macular edema. Front. Med. 2023, 10, 1128811. [Google Scholar] [CrossRef]
- Daruich, A.; Matet, A.; Moulin, A.; Kowalczuk, L.; Nicolas, M.; Sellam, A.; Rothschild, P.R.; Omri, S.; Gelize, E.; Jonet, L.; et al. Mechanisms of macular edema: Beyond the surface. Prog. Retin. Eye Res. 2018, 63, 20–68. [Google Scholar] [CrossRef]
- Carr, J.M.; Ashander, L.M.; Calvert, J.K.; Ma, Y.; Aloia, A.; Bracho, G.G.; Chee, S.P.; Appukuttan, B.; Smith, J.R. Molecular responses of human retinal cells to infection with dengue virus. Mediat. Inflamm. 2017, 2017, 3164375. [Google Scholar] [CrossRef]
- Soe, H.J.; Khan, A.M.; Manikam, R.; Samudi Raju, C.; Vanhoutte, P.; Sekaran, S.D. High dengue virus load differentially modulates human microvascular endothelial barrier function during early infection. J. Gen. Virol. 2017, 98, 2993–3007. [Google Scholar] [CrossRef]
- Syrbe, S.; Kuhrt, H.; Gartner, U.; Habermann, G.; Wiedemann, P.; Bringmann, A.; Reichenbach, A. Müller glial cells of the primate foveola: An electron microscopical study. Exp. Eye Res. 2018, 167, 110–117. [Google Scholar] [CrossRef]
- Reichenbach, A.; Bringmann, A. New functions of Müller cells. Glia 2013, 61, 651–678. [Google Scholar] [CrossRef]
- Toft-Kehler, A.K.; Skytt, D.M.; Kolko, M. A perspective on the Müller cell-neuron metabolic partnership in the inner retina. Mol. Neurobiol. 2018, 55, 5353–5361. [Google Scholar] [CrossRef]
- Vecino, E.; Rodriguez, F.D.; Ruzafa, N.; Pereiro, X.; Sharma, S.C. Glia-neuron interactions in the mammalian retina. Prog. Retin. Eye Res. 2016, 51, 1–40. [Google Scholar] [CrossRef]
- Xu, L.; Ruan, G.; Dai, H.; Liu, A.C.; Penn, J.; McMahon, D.G. Mammalian retinal Müller cells have circadian clock function. Mol. Vis. 2016, 22, 275–283. [Google Scholar]
- Drescher, K.M.; Whittum-Hudson, J.A. Evidence for induction of interferon-alpha and interferon-beta in retinal glial cells of Müller. Virology 1997, 234, 309–316. [Google Scholar] [CrossRef]
- Zhang, M.; Xin, H.; Atherton, S.S. Murine cytomegalovirus (MCMV) spreads to and replicates in the retina after endotoxin-induced disruption of the blood-retinal barrier of immunosuppressed BALB/c mice. J. Neurovirol. 2005, 11, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Luo, H.; Liu, H.; Ha, Y.; Mays, E.R.; Lawrence, R.E.; Winkelmann, E.; Barrett, A.D.; Smith, S.B.; Wang, M.; et al. p38MAPK plays a critical role in induction of a pro-inflammatory phenotype of retinal Müller cells following Zika virus infection. Antivir. Res. 2017, 145, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Rochet, E.; Appukuttan, B.; Ma, Y.; Ashander, L.M.; Smith, J.R. Expression of long non-coding RNAs by human retinal Müller glial cells infected with clonal and exotic virulent Toxoplasma gondii. Noncoding RNA 2019, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Geller, S.F.; Ge, P.S.; Visel, M.; Greenberg, K.P.; Flannery, J.G. Functional promoter testing using a modified lentiviral transfer vector. Mol. Vis. 2007, 13, 730–739. [Google Scholar] [PubMed]
- Klimczak, R.R.; Koerber, J.T.; Dalkara, D.; Flannery, J.G.; Schaffer, D.V. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Müller cells. PLoS ONE 2009, 4, e7467. [Google Scholar] [CrossRef]
- Kumar, A.; Pandey, R.K.; Miller, L.J.; Singh, P.K.; Kanwar, M. Müller glia in retinal innate immunity: A perspective on their roles in endophthalmitis. Crit. Rev. Immunol. 2013, 33, 119–135. [Google Scholar] [CrossRef]
- Kumar, A.; Shamsuddin, N. Retinal Müller glia initiate innate response to infectious stimuli via toll-like receptor signaling. PLoS ONE 2012, 7, e29830. [Google Scholar] [CrossRef]
- Mursalin, M.H.; Coburn, P.S.; Livingston, E.; Miller, F.C.; Astley, R.; Flores-Mireles, A.L.; Callegan, M.C. Bacillus S-layer-mediated innate interactions during endophthalmitis. Front. Immunol. 2020, 11, 215. [Google Scholar] [CrossRef]
- Limb, G.A.; Salt, T.E.; Munro, P.M.; Moss, S.E.; Khaw, P.T. In vitro characterization of a spontaneously immortalized human Müller cell line (MIO-M1). Invest. Ophthalmol. Vis. Sci. 2002, 43, 864–869. [Google Scholar]
- Dunn, K.C.; Aotaki-Keen, A.E.; Putkey, F.R.; Hjelmeland, L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996, 62, 155–169. [Google Scholar] [CrossRef]
- Smith, J.R.; Ashander, L.M.; Ma, Y.; Rochet, E.; Furtado, J.M. Model systems for studying mechanisms of ocular toxoplasmosis. Methods Mol. Biol. 2020, 2071, 297–321. [Google Scholar] [CrossRef]
- Pryor, M.J.; Carr, J.M.; Hocking, H.; Davidson, A.D.; Li, P.; Wright, P.J. Replication of dengue virus type 2 in human monocyte-derived macrophages: Comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am. J. Trop. Med. Hyg. 2001, 65, 427–434. [Google Scholar] [CrossRef]
- Wati, S.; Li, P.; Burrell, C.J.; Carr, J.M. Dengue virus (DV) replication in monocyte-derived macrophages is not affected by tumor necrosis factor alpha (TNF-alpha), and DV infection induces altered responsiveness to TNF-alpha stimulation. J. Virol. 2007, 81, 10161–10171. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Gualano, R.C.; Pryor, M.J.; Cauchi, M.R.; Wright, P.J.; Davidson, A.D. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 1998, 79, 437–446. [Google Scholar] [CrossRef]
- Kawali, A.; Mahendradas, P.; Mohan, A.; Mallavarapu, M.; Shetty, B. Epidemic retinitis. Ocul. Immunol. Inflamm. 2018, 27, 571–577. [Google Scholar] [CrossRef]
- Mi, H.; Ho, S.L.; Lim, W.K.; Wong, E.P.; Teoh, S.C. Trends in patterns of posterior uveitis and panuveitis in a tertiary institution in Singapore. Ocul. Immunol. Inflamm. 2015, 23, 329–338. [Google Scholar] [CrossRef]
- Janani, M.K.; Durgadevi, P.; Padmapriya, J.; Malathi, J.; Kulandai, L.T.; Rao Madhavan, H.N. First report on detection of dengue virus in the donor cornea. Cornea 2018, 37, 1586–1589. [Google Scholar] [CrossRef]
- Carod-Artal, F.J.; Wichmann, O.; Farrar, J.; Gascón, J. Neurological complications of dengue virus infection. Lancet Neurol. 2013, 12, 906–919. [Google Scholar] [CrossRef]
- Araújo, F.; Nogueira, R.; Araújo Mde, S.; Perdigão, A.; Cavalcanti, L.; Brilhante, R.; Rocha, M.; Vilar, D.F.; Holanda, S.S.; Braga Dde, M.; et al. Dengue in patients with central nervous system manifestations, Brazil. Emerg. Infect. Dis. 2012, 18, 677–679. [Google Scholar] [CrossRef]
- Vieira, M.; Costa, C.H.N.; Linhares, A.D.C.; Borba, A.S.; Henriques, D.F.; Silva, E.; Tavares, F.N.; Batista, F.M.A.; Guimarães, H.C.L.; Martins, L.C.; et al. Potential role of dengue virus, chikungunya virus and Zika virus in neurological diseases. Mem. Inst. Oswaldo Cruz 2018, 113, e170538. [Google Scholar] [CrossRef] [PubMed]
- Salomão, N.; Rabelo, K.; Basílio-de-Oliveira, C.; Basílio-de-Oliveira, R.; Geraldo, L.; Lima, F.; Dos Santos, F.; Nuovo, G.; Oliveira, E.R.A.; Paes, M. Fatal dengue cases reveal brain injury and viral replication in brain-resident cells associated with the local production of pro-inflammatory mediators. Viruses 2020, 12, 603. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.; Sanchez, G.; Pando, R.H.; Baquera, J.; Hernandez, D.; Mota, J.; Ramos, J.; Flores, A.; Llausas, E. Dengue virus in the brain of a fatal case of hemorrhagic dengue fever. J. Neurovirol. 1998, 4, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Norbury, A.J.; Calvert, J.K.; Al-Shujairi, W.H.; Cabezas-Falcon, S.; Tang, V.; Ong, L.C.; Alonso, S.L.; Smith, J.R.; Carr, J.M. Dengue virus infects the mouse eye following systemic or intracranial infection and induces inflammatory responses. J. Gen. Virol. 2020, 101, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Harris, E. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 2001, 289, 297–311. [Google Scholar] [CrossRef]
- Diamond, M.S.; Roberts, T.G.; Edgil, D.; Lu, B.; Ernst, J.; Harris, E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 2000, 74, 4957–4966. [Google Scholar] [CrossRef]
- Talarico, L.B.; Byrne, A.B.; Amarilla, S.; Lovera, D.; Vázquez, C.; Chamorro, G.; Acosta, P.L.; Ferretti, A.; Caballero, M.T.; Arbo, A.; et al. Characterization of type I interferon responses in dengue and severe dengue in children in Paraguay. J. Clin. Virol. 2017, 97, 10–17. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef]
- Ng, C.T.; Mendoza, J.L.; Garcia, K.C.; Oldstone, M.B. Alpha and beta type 1 interferon signaling: Passage for diverse biologic outcomes. Cell 2016, 164, 349–352. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Speranza, E.; Connor, J.H. Host transcriptional response to Ebola virus infection. Vaccines 2017, 5, 30. [Google Scholar] [CrossRef]
- Cowell, E.; Kris, L.P.; Bracho-Granado, G.; Jaber, H.; Smith, J.R.; Carr, J.M. Zika virus infection of retinal cells and the developing mouse eye induces host responses that contrasts to the brain and dengue virus infection. J. Neurovirol 2023, 29, 187–202. [Google Scholar] [CrossRef]
- Ashley, C.L.; Abendroth, A.; McSharry, B.P.; Slobedman, B. Interferon-independent innate responses to cytomegalovirus. Front. Immunol. 2019, 10, 2751. [Google Scholar] [CrossRef]
- Mlcochova, P.; Winstone, H.; Zuliani-Alvarez, L.; Gupta, R.K. TLR4-mediated pathway triggers interferon-independent G0 arrest and antiviral SAMHD1 activity in macrophages. Cell. Rep. 2020, 30, 3972–3980.e3975. [Google Scholar] [CrossRef]
- Lindqvist, R.; Kurhade, C.; Gilthorpe, J.D.; Överby, A.K. Cell-type- and region-specific restriction of neurotropic flavivirus infection by viperin. J. Neuroinflamm. 2018, 15, 80. [Google Scholar] [CrossRef]
- Dzimianski, J.V.; Scholte, F.E.M.; Bergeron, E.; Pegan, S.D. ISG15: It’s complicated. J. Mol. Biol. 2019, 431, 4203–4216. [Google Scholar] [CrossRef]
- Freitas, B.T.; Scholte, F.E.M.; Bergeron, É.; Pegan, S.D. How ISG15 combats viral infection. Virus Res. 2020, 286, 198036. [Google Scholar] [CrossRef]
- Helbig, K.J.; Beard, M.R. The role of viperin in the innate antiviral response. J. Mol. Biol. 2014, 426, 1210–1219. [Google Scholar] [CrossRef]
- Stirnweiss, A.; Ksienzyk, A.; Klages, K.; Rand, U.; Grashoff, M.; Hauser, H.; Kröger, A. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 2010, 184, 5179–5185. [Google Scholar] [CrossRef]
- Helbig, K.J.; Carr, J.M.; Calvert, J.K.; Wati, S.; Clarke, J.N.; Eyre, N.S.; Narayana, S.K.; Fiches, G.N.; McCartney, E.M.; Beard, M.R. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 2013, 7, e2178. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoek, K.H.; Eyre, N.S.; Shue, B.; Khantisitthiporn, O.; Glab-Ampi, K.; Carr, J.M.; Gartner, M.J.; Jolly, L.A.; Thomas, P.Q.; Adikusuma, F.; et al. Viperin is an important host restriction factor in control of Zika virus infection. Sci. Rep. 2017, 7, 4475. [Google Scholar] [CrossRef] [PubMed]
- Arias, J.; Valero, N.; Mosquera, J.; Montiel, M.; Reyes, E.; Larreal, Y.; Alvarez-Mon, M. Increased expression of cytokines, soluble cytokine receptors, soluble apoptosis ligand and apoptosis in dengue. Virology 2014, 452–453, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Tuyen, T.T.; Viet, N.T.; Hang, N.T.; Giang, N.T.; Anh, D.D.; Anh, D.T.; Hung, H.V.; Quyet, D.; Toan, N.L.; Cam, T.D.; et al. Pro-inflammatory cytokines are modulated in Vietnamese patients with dengue fever. Viral Immunol. 2020, 33, 514–520. [Google Scholar] [CrossRef]
- Ng, A.W.; Teoh, S.C. Dengue eye disease. Surv. Ophthalmol. 2015, 60, 106–114. [Google Scholar] [CrossRef]
- Teoh, S.C.; Chee, C.K.; Laude, A.; Goh, K.Y.; Barkham, T.; Ang, B.S.; Eye Institute Dengue-related Ophthalmic Complications Workgroup. Optical coherence tomography patterns as predictors of visual outcome in dengue-related maculopathy. Retina 2010, 30, 390–398. [Google Scholar] [CrossRef]
- Lim, W.K.; Mathur, R.; Koh, A.; Yeoh, R.; Chee, S.P. Ocular manifestations of dengue fever. Ophthalmology 2004, 111, 2057–2064. [Google Scholar] [CrossRef]
- Chang, P.E.; Cheng, C.L.; Asok, K.; Fong, K.Y.; Chee, S.P.; Tan, C.K. Visual disturbances in dengue fever: An answer at last? Singap. Med. J. 2007, 48, e71–e73. [Google Scholar]
- Tan, S.Y.; Kumar, G.; Surrun, S.K.; Ong, Y.Y. Dengue maculopathy: A case report. Travel. Med. Infect. Dis. 2007, 5, 62–63. [Google Scholar] [CrossRef]
- Gupta, S.; Das, D. Subhyaloid haemorrhage in dengue fever. J. Indian Med. Assoc. 2013, 111, 623–624. [Google Scholar]
- Yamamoto, K.; Takahashi, H.; Kanno, M.; Noda, Y.; Fujino, Y. Changes in parafoveal retinal thickness and subfoveal choroidal thickness in a patient with dengue fever-associated maculopathy. J. Ophthalmic Inflamm. Infect. 2013, 3, 63. [Google Scholar] [CrossRef]
- Li, M.; Zhang, X.; Ji, Y.; Ye, B.; Wen, F. Acute macular neuroretinopathy in dengue fever: Short-term prospectively followed up case series. JAMA Ophthalmol. 2015, 133, 1329–1333. [Google Scholar] [CrossRef]
- Ooi, K.G.; Inglis, H.; Paramanathan, N.; Downie, J.A.; Hennessy, M.P. Dengue fever-associated maculopathy and panuveitis in Australia. Case Rep. Ophthalmol. Med. 2016, 2016, 5704695. [Google Scholar] [CrossRef]
- Chee, E.; Sims, J.L.; Jap, A.; Tan, B.H.; Oh, H.; Chee, S.P. Comparison of prevalence of dengue maculopathy during two epidemics with differing predominant serotypes. Am. J. Ophthalmol. 2009, 148, 910–913. [Google Scholar] [CrossRef]
- Agarwal, A.; Aggarwal, K.; Dogra, M.; Kumar, A.; Akella, M.; Katoch, D.; Bansal, R.; Singh, R.; Gupta, V.; OCTA Study Group. Dengue-induced inflammatory, ischemic foveolitis and outer maculopathy: A swept-source imaging evaluation. Ophthalmol. Retina 2019, 3, 170–177. [Google Scholar] [CrossRef]
- Spaide, R.F. Retinal vascular cystoid macular edema: Review and new theory. Retina 2016, 36, 1823–1842. [Google Scholar] [CrossRef]
- Wang, X.; Wu, M.; Cao, Y.; Zhang, Z.; Guo, F.; Li, X.; Zhang, Y. Exploring the role of programmed cell death protein 1 and its ligand 1 in eye diseases. Crit. Rev. Clin. Lab. Sci. 2019, 56, 18–32. [Google Scholar] [CrossRef]
- Schönrich, G.; Raftery, M.J. The PD-1/PD-L1 axis and virus infections: A delicate balance. Front. Cell. Infect. Microbiol. 2019, 9, 207. [Google Scholar] [CrossRef]
- Jubel, J.M.; Barbati, Z.R.; Burger, C.; Wirtz, D.C.; Schildberg, F.A. The role of PD-1 in acute and chronic infection. Front. Immunol. 2020, 11, 487. [Google Scholar] [CrossRef]
- Nightingale, Z.D.; Patkar, C.; Rothman, A.L. Viral replication and paracrine effects result in distinct, functional responses of dendritic cells following infection with dengue 2 virus. J. Leukoc. Biol. 2008, 84, 1028–1038. [Google Scholar] [CrossRef]
- Lukowski, S.W.; Lo, C.Y.; Sharov, A.A.; Nguyen, Q.; Fang, L.; Hung, S.S.; Zhu, L.; Zhang, T.; Grunert, U.; Nguyen, T.; et al. A single-cell transcriptome atlas of the adult human retina. EMBO J. 2019, 38, e100811. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, H.; Chen, P.W.; Alizadeh, H.; He, Y.; Hogan, R.N.; Niederkorn, J.Y. PD-L1 expression on human ocular cells and its possible role in regulating immune-mediated ocular inflammation. Invest. Ophthalmol. Vis. Sci. 2009, 50, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Ashander, L.M.; Lumsden, A.L.; Dawson, A.C.; Ma, Y.; Ferreira, L.B.; Oliver, G.F.; Appukuttan, B.; Carr, J.M.; Smith, J.R. Infection of human retinal pigment epithelial cells with dengue virus strains isolated during outbreaks in Singapore. Microorganisms 2022, 10, 310. [Google Scholar] [CrossRef] [PubMed]

| Strain Name | GenBank ID | Serotype | Year of Isolation |
|---|---|---|---|
| EHI0393Y04 | EU069606.1 | DENV1 | 2004 |
| EHI0418Y05 | EU069594.1 | DENV1 | 2005 |
| EHI0377Y04 | JN851123.1 | DENV2 | 2004 |
| EHI0578Y05 | JN851126.1 | DENV2 | 2005 |
| EHI0043Y07 | GQ357691.1 | DENV1 | 2007 |
| EHI0169Y07 | GQ357690.1 | DENV1 | 2007 |
| Transcript | Primer Sequences | Expected Product Size (bp) | |
|---|---|---|---|
| DENV | FWD REV | 5′-CTA TGC TAA GCT TGA GCC CCG TC-3′ 5′-CGG ATC CTC TAG AAC CTG TTG-3′ | 400 |
| IFN-α | FWD REV | 5′-CAA GGT TCA GAG TCA CCC ATC-3′ 5′-CAG AGA GCA GCT TGA CTT GCA-3′ | 120 |
| IFN-β | FWD REV | 5′-AAA CTC ATG AGC AGT CTG CA-3′ 5′-AGG AGA TCT TCA GTT TCG GAG G-3′ | 168 |
| ISG15 | FWD REV | 5′-GAG AGG CAG CGA ACT CAT CT-3′ 5′-AGC ATC TTC ACC GTC AGG TC-3′ | 231 |
| RSAD2 | FWD REV | 5′-TGA CGG AAC AGA TCA AAG CA-3′ 5′-GCA CCA AGC AGG ACA CTT CT-3′ | 174 |
| TNF-α | FWD REV | 5′-TCT CGA ACC CCG AGT GAC AA-3′ 5′-TGA AGA GGA CCT GGG AGT AG-3′ | 482 |
| IL-1β | FWD REV | 5′-TGA CCT GAG CAC CTT CTT TC-3′ 5′-CAG CTG TAG AGT GGG CTT ATC-3′ | 331 |
| IL-6 | FWD REV | 5′-ATG AAC TCC TTC TCC ACA AGC GC-3′ 5′-GAA GAG CCC TCA GGC TGG ACT G-3′ | 628 |
| IL-10 | FWD REV | 5′-GTG ATG CCC CAA GCT GAG A-3′ 5′-GCA TCT GGC AAC CCT ACA ACA AG-3′ | 138 |
| PD-L1 | FWD REV | 5′-ACG CAT TTA CTG TCA CGG TTC C-3′ 5′-GAC TTC GGC CTT GGG GTA GC-3′ | 446 |
| PD-L2 | FWD REV | 5′-CAA CTT GGC TGC TTC ACA TTT T-3′ 5′-TGT GGT GAC AGG TCT TTT TGT TGT-3′ | 137 |
| FASL | FWD REV | 5′-GGG TGG ACT GGG GTG GCC TAT-3′ 5′-GGA TTG GGC CTG GGG ATG TTT CA-3′ | 126 |
| RPLP0 | FWD REV | 5′-GCA GCA TCT ACA ACC CTG AA-3′ 5′-GCA GAT GGA TCA GCC AAG AA-3′ | 235 |
| β-Actin | FWD REV | 5′-TCA AGA TCA TTG CTC CTC CTG AG-3′ 5′-ACA TCT GCT GGA AGG TGG ACA-3′ | 87 |
| GAPDH | FWD REV | 5′-AGC TGA ACG GGA AGC TCA CTG G-3′ 5′-GGA GTG GGT GTC GCT GTT GAA GTC-3′ | 209 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliver, G.F.; Ashander, L.M.; Dawson, A.C.; Ma, Y.; Carr, J.M.; Williams, K.A.; Smith, J.R. Dengue Virus Infection of Human Retinal Müller Glial Cells. Viruses 2023, 15, 1410. https://doi.org/10.3390/v15071410
Oliver GF, Ashander LM, Dawson AC, Ma Y, Carr JM, Williams KA, Smith JR. Dengue Virus Infection of Human Retinal Müller Glial Cells. Viruses. 2023; 15(7):1410. https://doi.org/10.3390/v15071410
Chicago/Turabian StyleOliver, Genevieve F., Liam M. Ashander, Abby C. Dawson, Yuefang Ma, Jillian M. Carr, Keryn A. Williams, and Justine R. Smith. 2023. "Dengue Virus Infection of Human Retinal Müller Glial Cells" Viruses 15, no. 7: 1410. https://doi.org/10.3390/v15071410
APA StyleOliver, G. F., Ashander, L. M., Dawson, A. C., Ma, Y., Carr, J. M., Williams, K. A., & Smith, J. R. (2023). Dengue Virus Infection of Human Retinal Müller Glial Cells. Viruses, 15(7), 1410. https://doi.org/10.3390/v15071410






