Pellino Proteins in Viral Immunity and Pathogenesis
Abstract
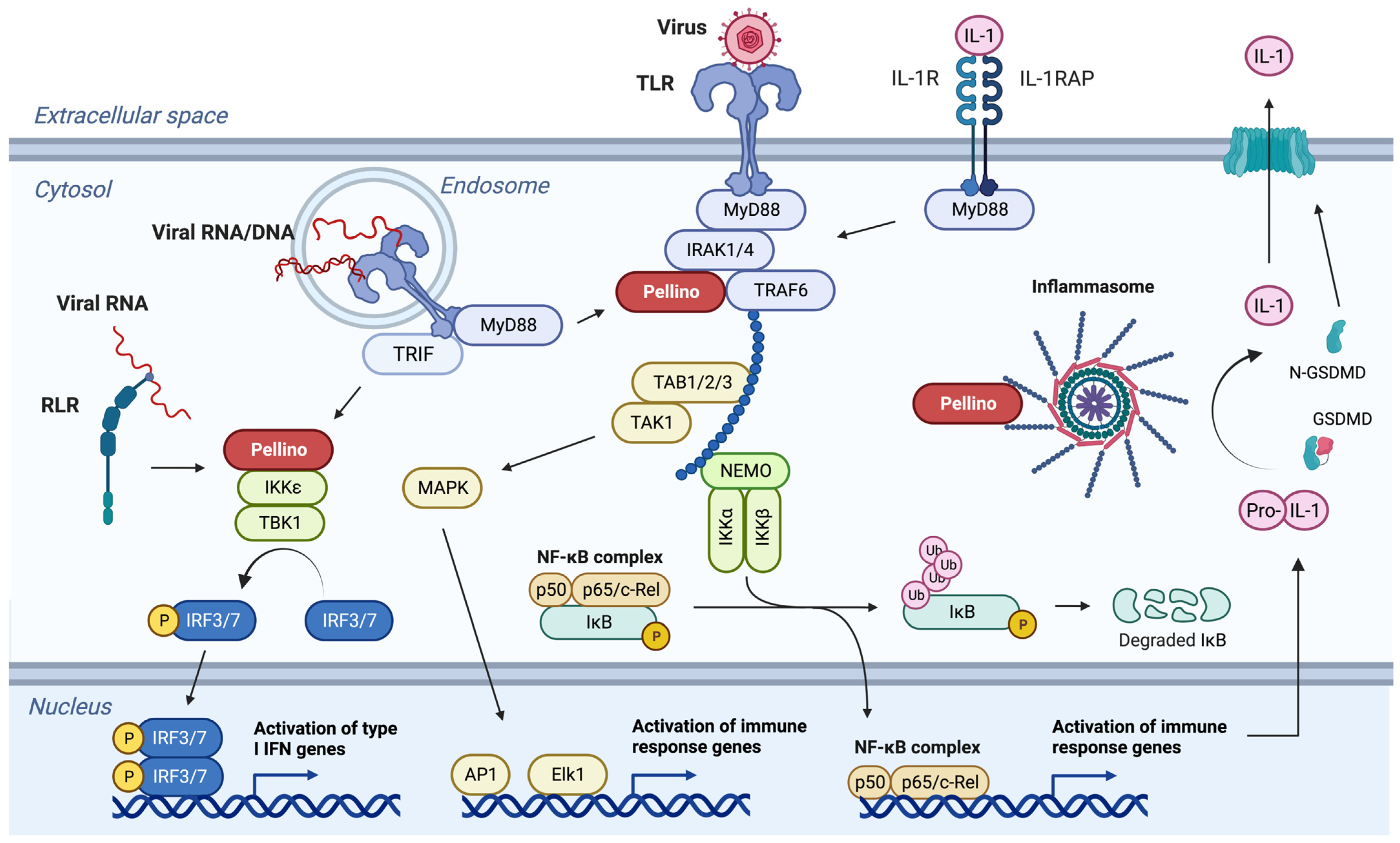
:1. Introduction
2. Initiation of Immune Responses
3. Viral Immune Evasion
4. Excess Inflammation and Pathogenesis
5. Immunoregulatory Functions
6. Regulation of Cell Death Mechanisms
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: Styles, structures and functions. Mol. Biomed. 2021, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Grosshans, J.; Schnorrer, F.; Nusslein-Volhard, C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech. Dev. 1999, 81, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Li, X. The emerging roles of Pellino family in pattern recognition receptor signaling. Front. Immunol. 2022, 13, 728794. [Google Scholar] [CrossRef] [PubMed]
- Humphries, F.; Moynagh, P.N. Molecular and physiological roles of Pellino E3 ubiquitin ligases in immunity. Immunol. Rev. 2015, 266, 93–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauvliege, R.; Janssens, S.; Beyaert, R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: A role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006, 580, 4697–4702. [Google Scholar] [CrossRef]
- Butler, M.P.; Hanly, J.A.; Moynagh, P.N. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: Direct evidence for Pellino proteins being ubiquitin-protein isopeptide ligases. J. Biol. Chem. 2007, 282, 29729–29737. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Huoh, Y.S.; Schmitz, K.R.; Jensen, L.E.; Ferguson, K.M. Pellino proteins contain a cryptic FHA domain that mediates interaction with phosphorylated IRAK1. Structure 2008, 161, 1806–1816. [Google Scholar] [CrossRef] [Green Version]
- Rich, T.; Allen, R.L.; Lucas, A.-M.; Stewart, A.; Trowsdale, J. Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics 2000, 52, 145–149. [Google Scholar] [CrossRef]
- Cluxton, C.D.; Caffrey, B.E.; Kinsella, G.K.; Moynagh, P.N.; Fares, M.A.; Fallon, P.G. Functional conservation of an ancestral Pellino protein in helminth species. Sci. Rep. 2015, 5, 11687. [Google Scholar] [CrossRef] [Green Version]
- Ravichandran, G.; Kumaresan, V.; Arasu, M.V.; Al-Dhabi, N.A.; Ganesh, M.R.; Mahesh, A.; Dhayalan, A.; Pasupuleti, M.; Arockiaraj, J. Pellino-1 derived cationic antimicrobial prawn peptide: Bactericidal activity, toxicity and mode of action. Mol. Immunol. 2016, 78, 171–182. [Google Scholar] [CrossRef]
- Li, C.; Chai, J.; Li, H.; Zuo, H.; Wang, S.; Qiu, W.; Weng, S.; He, J.; Xu, X. Pellino protein from pacific white shrimp Litopenaeus vannamei positively regulates NF-κB activation. Dev. Comp. Immunol. 2014, 44, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cheng, W.; Zheng, J.; Wang, P.; Liu, Q.; Li, Z.; Shi, T.; Zhou, Y.; Mao, Y.; Yu, X. Identification and molecular characterization of a Pellino protein in kuruma prawn (Marsupenaeus japonicus) in response to white spot syndrome virus and vibrio parahaemolyticus infection. Int. J. Mol. Sci. 2020, 21, 1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, L.E.; Whitehead, A.S. Pellino3, a novel member of the Pellino protein family, promotes activation of c-Jun and Elk-1 and may act as a scaffolding protein. J. Immunol. 2003, 171, 1500–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphries, F.; Bergin, R.; Jackson, R.; Delagic, N.; Wang, B.; Yang, S.; Dubois, A.V.; Ingram, R.J.; Moynagh, P.N. The E3 ubiquitin ligase Pellino2 mediates priming of the NLRP3 inflammasome. Nat. Commun. 2018, 9, 1560. [Google Scholar] [CrossRef] [Green Version]
- Siednienko, J.; Jackson, R.; Mellett, M.; Delagic, N.; Yang, S.; Wang, B.; Tang, L.S.; Callanan, J.J.; Mahon, B.P.; Moynagh, P.N. Pellino3 targets the IRF7 pathway and facilitates autoregulation of TLR3- and viral-induced expression of type I interferons. Nat. Immunol. 2012, 13, 1055–1062. [Google Scholar] [CrossRef]
- Cai, K.Q.; Shellhamer, C.; Akiyama, T.; Jensen, L.E. Pellino1 restricts herpes simplex virus infections in the epidermis and dissemination to sebaceous glands. J. Investig. Dermatol. 2023, 143, 639–647. [Google Scholar] [CrossRef]
- Chang, M.; Jin, W.; Sun, S.C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 2009, 10, 1089–1095. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Winkelmann, E.R.; Zhu, S.; Ru, W.; Mays, E.; Silvas, J.A.; Vollmer, L.L.; Gao, J.; Peng, B.H.; Bopp, N.E.; et al. Peli1 facilitates virus replication and promotes neuroinflammation during West Nile virus infection. J. Clin. Investig. 2018, 128, 4980–4991. [Google Scholar] [CrossRef]
- Hatton, A.A.; Guerra, F.E. Scratching the surface takes a toll: Immune recognition of viral proteins by surface Toll-like receptors. Viruses 2022, 15, 52. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Chou, W.C.; Jha, S.; Linhoff, M.W.; Ting, J.P. The NLR gene family: From discovery to present day. Nat. Rev. Immunol. 2023. [Google Scholar] [CrossRef]
- Bennett, J.A.; Prince, L.R.; Parker, L.C.; Stokes, C.A.; de Bruin, H.G.; van den Berge, M.; Heijink, I.H.; Whyte, M.K.; Sabroe, I. Pellino-1 selectively regulates epithelial cell responses to rhinovirus. J. Virol. 2012, 86, 6595–6604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enesa, K.; Ordureau, A.; Smith, H.; Barford, D.; Cheung, P.C.F.; Patterson-Kane, J.; Arthur, J.S.C.; Cohen, P. Pellino1 Is required for interferon production by viral double-stranded RNA. J. Biol. Chem. 2012, 287, 34825–34835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ordureau, A.; Enesa, K.; Nanda, S.; Le Francois, B.; Peggie, M.; Prescott, A.; Albert, P.R.; Cohen, P. DEAF1 is a Pellino1-interacting protein required for interferon production by Sendai virus and double-stranded RNA. J. Biol. Chem. 2013, 288, 24569–24580. [Google Scholar] [CrossRef] [Green Version]
- Reniewicz, P.; Kula, A.; Makuch, E.; Ochnik, M.; Lipiński, T.; Siednienko, J. Ligase Pellino3 regulates macrophage action and survival in response to VSV infection in RIG-I-dependent path. Oxid. Med. Cell Longev. 2021, 2021, 6668463. [Google Scholar] [CrossRef] [PubMed]
- Marsh, E.K.; Prestwich, E.C.; Williams, L.; Hart, A.R.; Muir, C.F.; Parker, L.C.; Jonker, M.R.; Heijink, I.H.; Timens, W.; Fife, M.; et al. Pellino-1 regulates the responses of the airway to viral infection. Front. Cell Infect. Microbiol. 2020, 10, 456. [Google Scholar] [CrossRef]
- Olaru, F.; Jensen, L.E. Staphylococcus aureus stimulates neutrophil targeting chemokine expression in keratinocytes through an autocrine IL-1α signaling loop. J. Investig. Dermatol. 2010, 130, 1866–1876. [Google Scholar] [CrossRef] [Green Version]
- Marrakchi, S.; Guigue, P.; Renshaw, B.R.; Puel, A.; Pei, X.-Y.; Fraitag, S.; Zribi, J.; Bal, E.; Cluzeau, C.; Chrabieh, M.; et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N. Engl. J. Med. 2011, 365, 620–628. [Google Scholar] [CrossRef]
- Wojtasiak, M.; Pickett, D.L.; Tate, M.D.; Bedoui, S.; Job, E.R.; Whitney, P.G.; Brooks, A.G.; Reading, P.C. Gr-1+ cells, but not neutrophils, limit virus replication and lesion development following flank infection of mice with herpes simplex virus type-1. Virology 2010, 407, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Hor, J.L.; Heath, W.R.; Mueller, S.N. Neutrophils are dispensable in the modulation of T cell immunity against cutaneous HSV-1 infection. Sci. Rep. 2017, 7, 41091. [Google Scholar] [CrossRef]
- Gardner, J.K.; Swaims-Kohlmeier, A.; Herbst-Kralovetz, M.M. IL-36γ is a key regulator of neutrophil infiltration in the vaginal microenvironment and limits neuroinvasion in genital HSV-2 infection. J. Immunol. 2019, 203, 2655–2664. [Google Scholar] [CrossRef]
- Peperzak, V.; Veraar, E.A.; Xiao, Y.; Babala, N.; Thiadens, K.; Brugmans, M.; Borst, J. CD8+ T cells produce the chemokine CXCL10 in response to CD27/CD70 costimulation to promote generation of the CD8+ effector T cell pool. J. Immunol. 2013, 191, 3025–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozga, A.J.; Chow, M.T.; Lopes, M.E.; Servis, R.L.; Di Pilato, M.; Dehio, P.; Lian, J.; Mempel, T.R.; Luster, A.D. CXCL10 chemokine regulates heterogeneity of the CD8(+) T cell response and viral set point during chronic infection. Immunity 2022, 55, 82–97.e8. [Google Scholar] [CrossRef] [PubMed]
- Ocón, B.; Pan, J.; Dinh, T.T.; Chen, W.; Ballet, R.; Bscheider, M.; Habtezion, A.; Tu, H.; Zabel, B.A.; Butcher, E.C. A mucosal and cutaneous chemokine ligand for the lymphocyte chemoattractant receptor GPR15. Front. Immunol. 2017, 8, 1111. [Google Scholar] [CrossRef] [Green Version]
- Suply, T.; Hannedouche, S.; Carte, N.; Li, J.; Grosshans, B.; Schaefer, M.; Raad, L.; Beck, V.; Vidal, S.; Hiou-Feige, A.; et al. A natural ligand for the orphan receptor GPR15 modulates lymphocyte recruitment to epithelia. Sci. Signal. 2017, 10, eaal0180. [Google Scholar] [CrossRef] [PubMed]
- Dainichi, T.; Nakano, Y.; Doi, H.; Nakamizo, S.; Nakajima, S.; Matsumoto, R.; Farkas, T.; Wong, P.M.; Narang, V.; Moreno Traspas, R.; et al. C10orf99/GPR15L regulates proinflammatory response of keratinocytes and barrier formation of the skin. Front. Immunol. 2022, 13, 825032. [Google Scholar] [CrossRef]
- Jiang, Q.; Zhu, Z.; Mao, X. Ubiquitination is a major modulator for the activation of inflammasomes and pyroptosis. Biochim. Biophys. Acta Gene Regul. Mech. 2023, 194955. [Google Scholar] [CrossRef]
- Jensen, L.E. Interleukin-36 cytokines may overcome microbial immune evasion strategies that inhibit interleukin-1 family signaling. Sci. Signal. 2017, 10, eaan3589. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ko, C.J.; Li, Y.; Jie, Z.; Zhu, L.; Zhou, X.; Xie, X.; Gao, T.; Liu, T.; Cheng, X.; et al. Peli1 facilitates NLRP3 inflammasome activation by mediating ASC ubiquitination. Cell Rep. 2021, 37, 109904. [Google Scholar] [CrossRef]
- Liu, X.; Lin, Z.; Yin, X. Pellino2 accelerate inflammation and pyroptosis via the ubiquitination and activation of NLRP3 inflammation in model of pediatric pneumonia. Int. Immunopharmacol. 2022, 110, 108993. [Google Scholar] [CrossRef]
- Cristea, I.; Bruland, O.; Rødahl, E.; Bredrup, C. K(+) regulates relocation of Pellino-2 to the site of NLRP3 inflammasome activation in macrophages. FEBS Lett. 2021, 595, 2437–2446. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.; Bowie, A.G. Recent insights into innate immune nucleic acid sensing during viral infection. Curr. Opin. Immunol. 2022, 78, 102250. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; Borg, N.A. The multifaceted roles of NLRP3-modulating proteins in virus infection. Front. Immunol. 2022, 13, 987453. [Google Scholar] [CrossRef] [PubMed]
- Verdonck, S.; Nemegeer, J.; Vandenabeele, P.; Maelfait, J. Viral manipulation of host cell necroptosis and pyroptosis. Trends Microbiol. 2022, 30, 593–605. [Google Scholar] [CrossRef]
- Long, S.; Yang, L.; Dang, W.; Xin, S.; Jiang, M.; Zhang, W.; Li, J.; Wang, Y.; Zhang, S.; Lu, J. Cellular deubiquitylating enzyme: A regulatory factor of antiviral innate immunity. Front. Microbiol. 2021, 12, 805223. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, D.; Tian, Y.; Wang, B.; Zhu, F.; Yin, Q. Structural basis of higher order oligomerization of KSHV inhibitor of cGAS. Proc. Natl. Acad. Sci. USA 2022, 119, e2200285119. [Google Scholar] [CrossRef]
- Beachboard, D.C.; Horner, S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 2016, 32, 113–119. [Google Scholar] [CrossRef]
- Bhowmik, D.; Du, M.; Tian, Y.; Ma, S.; Wu, J.; Chen, Z.; Yin, Q.; Zhu, F. Cooperative DNA binding mediated by KicGAS/ORF52 oligomerization allows inhibition of DNA-induced phase separation and activation of cGAS. Nucleic Acids Res. 2021, 49, 9389–9403. [Google Scholar] [CrossRef]
- Milora, K.A.; Miller, S.L.; Sanmiguel, J.C.; Jensen, L.E. Interleukin-1α released from HSV-1 infected keratinocytes acts as a functional alarmin in the skin. Nat. Commun. 2014, 5, 5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, B.D.; Mellett, M.; Campos-Torres, A.; Kinsella, G.K.; Wang, B.; Moynagh, P.N. A poxviral homolog of the Pellino protein inhibits Toll and Toll-like receptor signalling. Eur. J. Immunol. 2011, 41, 798–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonso, C.L.; Tulman, E.R.; Lu, Z.; Oma, E.; Kutish, G.F.; Rock, D.L. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 1999, 73, 533–552. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, M.; Singh, S.K. Japanese Encephalitis Virus exploits microRNA-155 to suppress the non-canonical NF-κB pathway in human microglial cells. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194639. [Google Scholar] [CrossRef]
- Liu, W.H.; Kang, S.G.; Huang, Z.; Wu, C.J.; Jin, H.Y.; Maine, C.J.; Liu, Y.; Shepherd, J.; Sabouri-Ghomi, M.; Gonzalez-Martin, A.; et al. A miR-155-Peli1-c-Rel pathway controls the generation and function of T follicular helper cells. J. Exp. Med. 2016, 213, 1901–1919. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Li, G.; Wang, B.; Tian, B.; Gao, J.; Zou, J.; Shi, S.; Zhu, S.; Peng, B.H.; Adam, A.; et al. Peli1 signaling blockade attenuates congenital zika syndrome. PLoS Pathog. 2020, 16, e1008538. [Google Scholar] [CrossRef]
- Baines, K.J.; Hsu, A.C.; Tooze, M.; Gunawardhana, L.P.; Gibson, P.G.; Wark, P.A. Novel immune genes associated with excessive inflammatory and antiviral responses to rhinovirus in COPD. Respir. Res. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, D.; Ganesan, S.; Comstock, A.T.; Meldrum, C.A.; Mahidhara, R.; Goldsmith, A.M.; Curtis, J.L.; Martinez, F.J.; Hershenson, M.B.; Sajjan, U. Increased cytokine response of rhinovirus-infected airway epithelial cells in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2010, 182, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Tzieply, N.; Kuhn, A.-M.; Morbitzer, D.; Namgaladze, D.; Heeg, A.; Schaefer, L.; von Knethen, A.; Jensen, L.E.; Brüne, B. OxLDL inhibits LPS-induced IFNbeta expression by Pellino3- and IRAK1/4-dependent modification of TANK. Cell. Signal. 2012, 24, 1141–1149. [Google Scholar] [CrossRef]
- Kula, A.; Makuch, E.; Lisowska, M.; Reniewicz, P.; Lipiński, T.; Siednienko, J. Pellino3 ligase negatively regulates influenza B dependent RIG-I signalling through downregulation of TRAF3-mediated induction of the transcription factor IRF3 and IFNβ production. Immunology 2023, 169, 369–383. [Google Scholar] [CrossRef]
- Xiao, Y.; Jin, J.; Chang, M.; Chang, J.H.; Hu, H.; Zhou, X.; Brittain, G.C.; Stansberg, C.; Torkildsen, Ø.; Wang, X.; et al. Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat. Med. 2013, 19, 595–602. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Jin, J.; Zou, Q.; Hu, H.; Cheng, X.; Sun, S.C. Peli1 negatively regulates type I interferon induction and antiviral immunity in the CNS. Cell Biosci. 2015, 5, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.; Jin, W.; Chang, J.-H.; Xiao, Y.; Brittain, G.C.; Yu, J.; Zhou, X.; Wang, Y.-H.; Cheng, X.; Li, P.; et al. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat. Immunol. 2011, 12, 1002–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.C.; Lee, Y.S.; Lim, S.; Choi, H.K.; Lee, C.H.; Lee, E.K.; Hong, S.; Kim, I.H.; Kim, S.J.; Park, S.H. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat. Immunol. 2006, 7, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, J.H.; Kim, S.T.; Kwon, J.Y.; Hong, S.; Kim, S.J.; Park, S.H. Smad7 and Smad6 bind to discrete regions of Pellino-1 via their MH2 domains to mediate TGF-beta1-induced negative regulation of IL-1R/TLR signaling. Biochem. Biophys. Res. Commun. 2010, 393, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Mellor, A.L.; Lemos, H.; Huang, L. Indoleamine 2,3-dioxygenase and tolerance: Where are we now? Front. Immunol. 2017, 8, 1360. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, T.D.; Gerondakis, S. The c-Rel transcription factor in development and disease. Genes Cancer 2011, 2, 695–711. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Chang, M.; Sun, S.C. Peli: A family of signal-responsive E3 ubiquitin ligases mediating TLR signaling and T-cell tolerance. Cell. Mol. Immunol. 2012, 9, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Bigley, T.M.; Yang, L.; Kang, L.-I.; Saenz, J.B.; Victorino, F.; Yokoyama, W.M. Disruption of thymic central tolerance by infection with murine roseolovirus induces autoimmune gastritis. J. Exp. Med. 2022, 219, e20211403. [Google Scholar] [CrossRef]
- Smatti, M.K.; Cyprian, F.S.; Nasrallah, G.K.; Al Thani, A.A.; Almishal, R.O.; Yassine, H.M. Viruses and autoimmunity: A review on the potential interaction and molecular mechanisms. Viruses 2019, 11, 762. [Google Scholar] [CrossRef] [Green Version]
- Miyazawa, K.; Miyazono, K. Regulation of TGF-β family signaling by inhibitory Smads. Cold Spring. Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Fox, J.M.; Sage, L.K.; Poore, S.; Johnson, S.; Tompkins, S.M.; Tripp, R.A. Drug analog inhibition of indoleamine 2,3-dioxygenase (IDO) activity modifies pattern recognition receptor expression and proinflammatory cytokine responses early during influenza virus infection. J. Leukoc. Biol. 2014, 96, 447–452. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Aglietti, R.A.; Estevez, A.; Gupta, A.; Ramirez, M.G.; Liu, P.S.; Kayagaki, N.; Ciferri, C.; Dixit, V.M.; Dueber, E.C. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 2016, 113, 7858–7863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; He, W.-t.; Hu, L.; Li, J.; Fang, Y.; Wang, X.; Xu, X.; Wang, Z.; Huang, K.; Han, J. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 2016, 26, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Russo, H.M.; Rathkey, J.; Boyd-Tressler, A.; Katsnelson, M.A.; Abbott, D.W.; Dubyak, G.R. Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J. Immunol. 2016, 197, 1353–1367. [Google Scholar] [CrossRef] [Green Version]
- Sborgi, L.; Rühl, S.; Mulvihill, E.; Pipercevic, J.; Heilig, R.; Stahlberg, H.; Farady, C.J.; Müller, D.J.; Broz, P.; Hiller, S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016, 35, 1766–1778. [Google Scholar] [CrossRef]
- Pearson, J.S.; Murphy, J.M. Down the rabbit hole: Is necroptosis truly an innate response to infection? Cell Microbiol. 2017, 19. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.W.; Park, H.H.; Kim, S.; Chung, J.M.; Noh, H.J.; Kim, S.K.; Song, H.K.; Lee, C.W.; Morgan, M.J.; Kang, H.C.; et al. PELI1 selectively targets kinase-active RIP3 for ubiquitylation-dependent proteasomal degradation. Mol. Cell 2018, 70, 920–935.e7. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Meng, H.; Li, X.; Zhu, K.; Dong, K.; Mookhtiar, A.K.; Wei, H.; Li, Y.; Sun, S.C.; Yuan, J. PELI1 functions as a dual modulator of necroptosis and apoptosis by regulating ubiquitination of RIPK1 and mRNA levels of c-FLIP. Proc. Natl. Acad. Sci. USA 2017, 114, 11944–11949. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, L.; Mostowy, S.; Sancho-Shimizu, V. Autophagy-virus interplay: From cell biology to human disease. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, K.; Niu, F.; Hu, G.; Guo, M.L.; Sil, S.; Buch, S. HIV Tat-mediated induction of autophagy regulates the disruption of ZO-1 in brain endothelial cells. Tissue Barriers 2020, 8, 1748983. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Pellino Protein | Downstream Degraded Target | Upstream Regulator |
|---|---|---|
| Pellino3 | TRAF6 [15] | |
| Pellino3 | TRAF3 [58] | |
| Pellino3 | TANK [57] | |
| Pellino1 | TRAF3 [59,60] | |
| Pellino1 | c-Rel [61] | miR-155 [53] |
| Pellino1 | Smad6 [62] | |
| Pellino1 | Smad7 [63] | |
| Pellino1 | IDO [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jensen, L.E. Pellino Proteins in Viral Immunity and Pathogenesis. Viruses 2023, 15, 1422. https://doi.org/10.3390/v15071422
Jensen LE. Pellino Proteins in Viral Immunity and Pathogenesis. Viruses. 2023; 15(7):1422. https://doi.org/10.3390/v15071422
Chicago/Turabian StyleJensen, Liselotte E. 2023. "Pellino Proteins in Viral Immunity and Pathogenesis" Viruses 15, no. 7: 1422. https://doi.org/10.3390/v15071422





