Cluster Analysis Identifies Distinct Patterns of T-Cell and Humoral Immune Responses Evolution Following a Third Dose of SARS-CoV-2 Vaccine in People Living with HIV
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Procedures
2.2.1. Quantification of Total SARS-CoV-2 Anti-Nucleocapsid Total Antibodies
2.2.2. Quantification of Total Anti-Spike IgG Antibodies
2.2.3. Quantification of SARS-CoV-2 Neutralising Antibodies
2.2.4. QuantiFERON SARS-CoV-2 Interferon-ɣ Release Assay
2.3. Statistical Analysis
3. Results
3.1. Baseline Study Cohort Characteristics of PLWH and HCWs
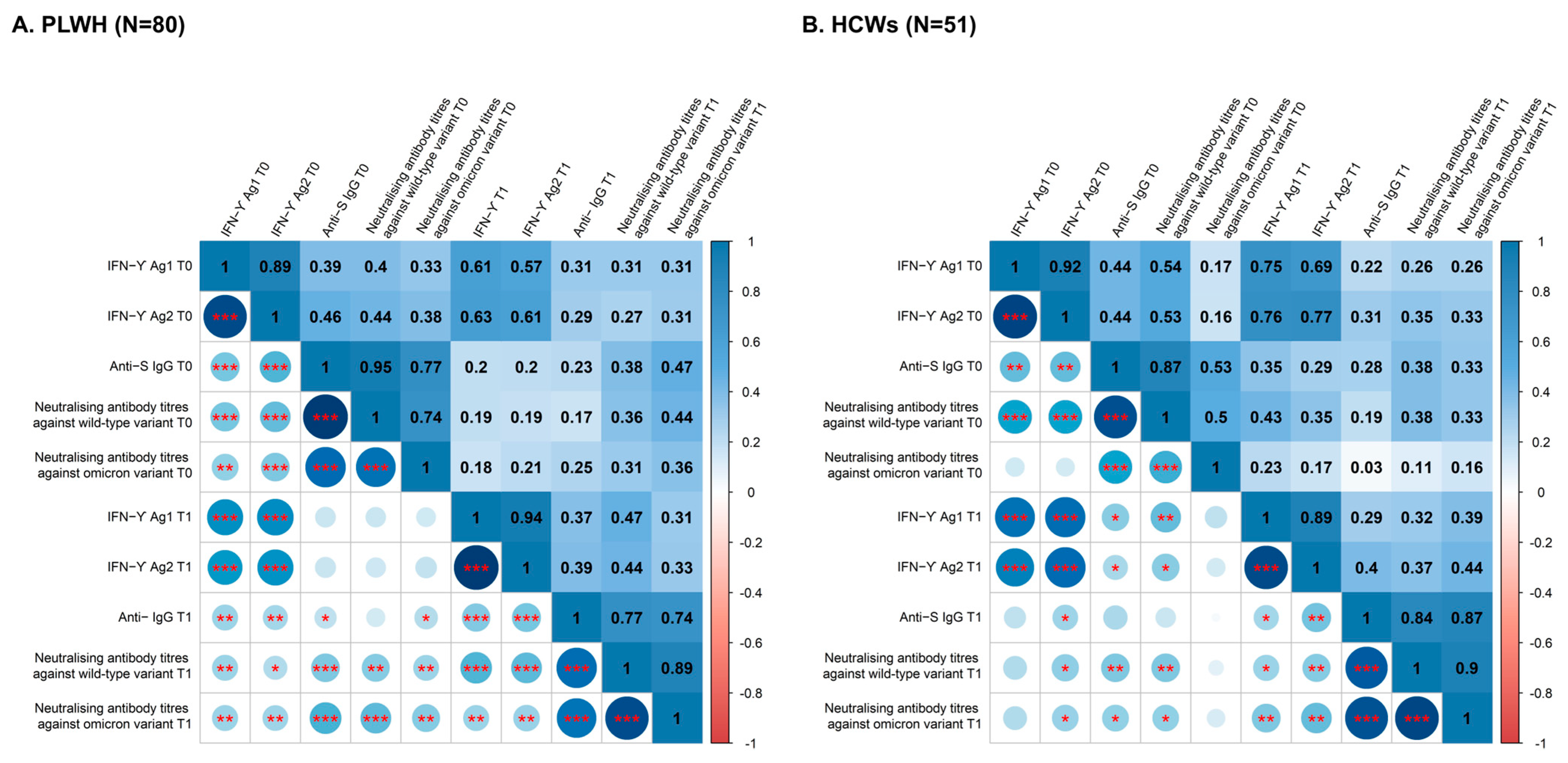
3.2. Relationship between T-Cell and Humoral Immune Responses
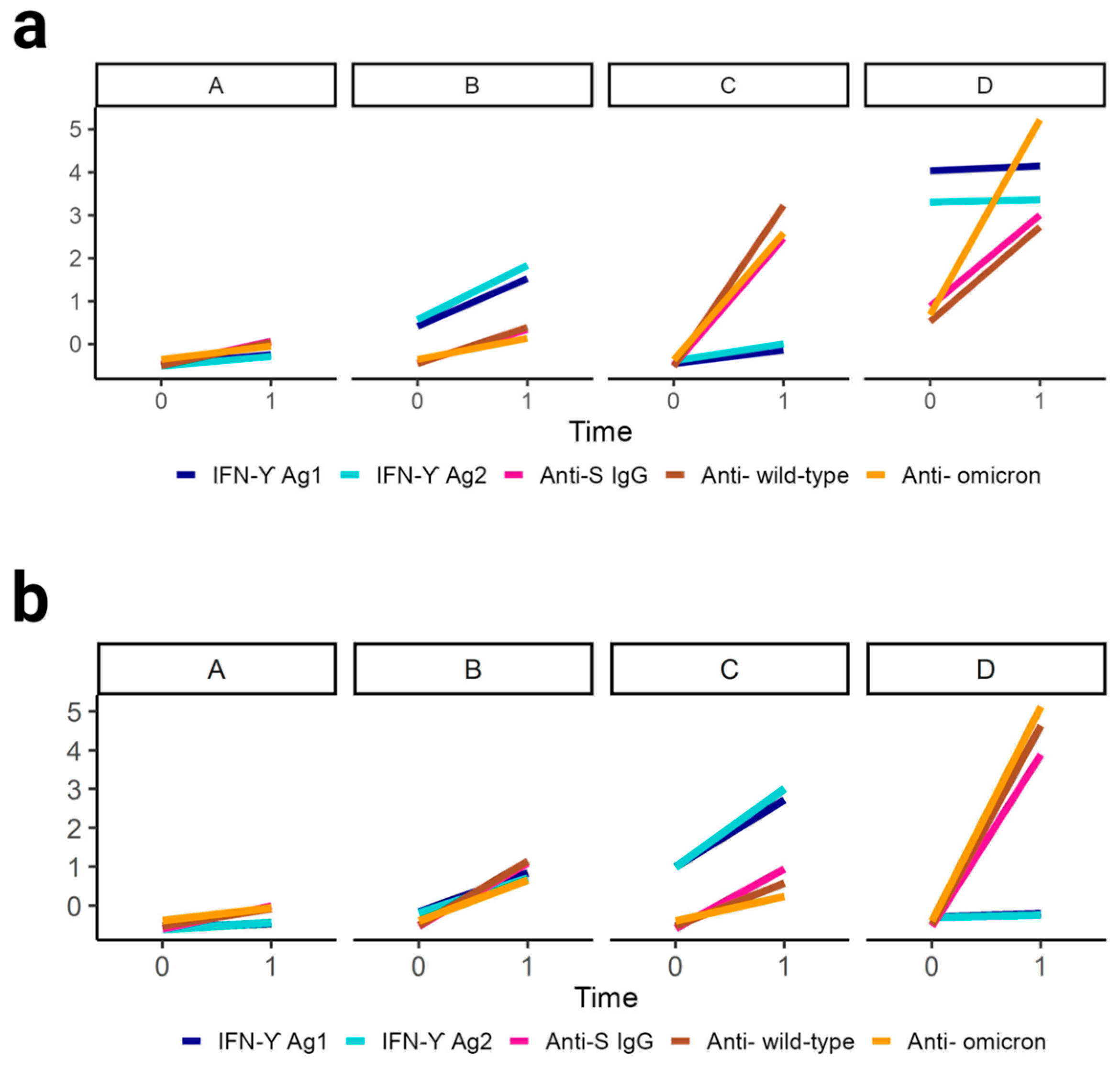
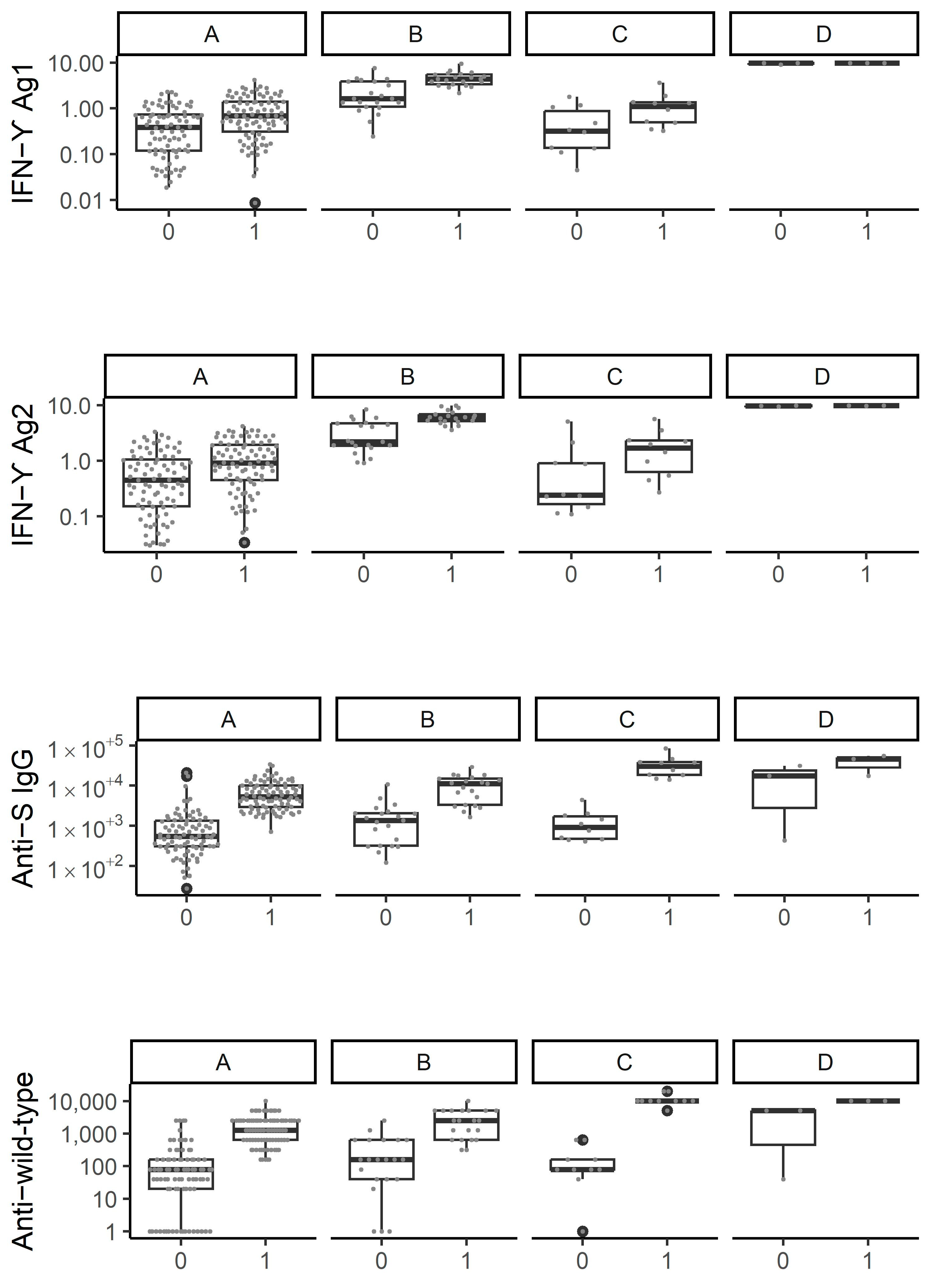
3.3. Cluster Analysis Identifies Four Distinct Patterns of Immune Response Evolution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Moss, P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022, 23, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Volger, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef]
- Chen, X.; Pan, Z.; Yue, S.; Yu, F.; Zhang, J.; Yang, Y.; Li, R.; Liu, B.; Yang, X.; Gao, L.; et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct. Target. Ther. 2020, 5, 180. [Google Scholar] [CrossRef] [PubMed]
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef]
- Liu, J.; Chandrashekar, A.; Sellers, D.; Barrett, J.; Jacob-Dolan, C.; Lifton, M.; McMahan, K.; Sciacca, M.; VanWyk, H.; Wu, C.; et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 2020, 603, 493–496. [Google Scholar] [CrossRef]
- Duly, K.; Farraye, F.A.; Bhat, S. COVID-19 vaccine use in immunocompromised patients: A commentary on evidence and recommendations. Am. J. Health Pharm. 2022, 79, 63–71. [Google Scholar] [CrossRef]
- Oyaert, M.; De Scheerder, M.A.; Van Herrewege, S.; Laureys, G.; Van Assche, S.; Cambron, M.; Naesens, L.; Hoste, L.; Claes, K.; Haerynck, F.; et al. Evaluation of Humoral and Cellular Responses in SARS-CoV-2 mRNA Vaccinated Immunocompromised Patients. Front. Immunol. 2022, 13, 858399. [Google Scholar] [CrossRef]
- El Moussaoui, M.E.; Desmecht, S.; Tashkeev, A.; Lambert, N.; Maes, N.; Braghini, J.; Marechal, N.; Quintana, C.; Briquet, K.; Gofflot, S.; et al. Reduced T-cell response following a third dose of SARS-CoV-2 vaccine in infection-naïve people living with HIV. J. Infect. 2022, 85, 702–769. [Google Scholar] [CrossRef]
- Grégoire, C.; Huynen, P.; Gofflot, S.; Seidel, L.; Maes, N.; Vranken, L.; Delcour, S.; Moutschen, M.; Hayette, M.-P.; Kolh, P.; et al. Predictive factors for the presence and long-term persistence of SARS-CoV-2 antibodies in healthcare and university workers. Sci. Rep. 2022, 12, 9790. [Google Scholar] [CrossRef] [PubMed]
- Nayrac, M.; Dubé, M.; Sannier, G.; Nicolas, A.; Marchitto, L.; Tastet, O.; Tauzin, A.; Brassard, N.; Lima-Barbosa, R.; Beaudoin-Bussières, G.; et al. Temporal associations of B and T cell immunity with robust vaccine responsiveness in a 16-week interval BNT162b2 regimen. Cell Rep. 2022, 39, 111013. [Google Scholar] [CrossRef]
- Goel, R.R.; Painter, M.M.; Apostolidis, S.A.; Mathew, D.; Meng, W.; Rosenfeld, A.M.; Lundgreen, K.A.; Reynaldi, A.; Khoury, D.S.; Pattekar, A.; et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 2021, 374, abm0829. [Google Scholar] [CrossRef] [PubMed]
- Painter, M.M.; Mathew, D.; Goel, R.R.; Apostolidis, S.A.; Pattekar, A.; Kuthuru, O.; Baxter, A.E.; Herati, R.S.; Oldridge, D.A.; Gouma, S.; et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 2021, 54, 2133–2142.e3. [Google Scholar] [CrossRef] [PubMed]
- Genolini, C.; Pingault, J.B.; Driss, T.; Côté, S.; Tremblay, R.; Vitaro, F.; Arnaud, C.; Falissard, B. KmL3D: A non-parametric algorithm for clustering joint trajectories. Comput. Methods Programs Biomed. 2013, 109, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Genolini, C.; Alacoque, X.; Sentenac, M.; Arnaud, C. Kml and kml3d: R Packages to Cluster Longitudinal Data. J. Stat. Softw. 2015, 65, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Menges, D.; Zens, K.D.; Balouz, T.; Caduff, N.; Llanas-Cornejo, D.; Aschmann, H.E.; Domenghino, A.; Pellaton, C.; Perreau, M.; Fenwick, C.; et al. Heterogenous humoral and cellular immune responses with distinct trajectories post-SARS-CoV-2 infection in a population-based cohort. Nat. Commun. 2022, 13, 4855. [Google Scholar] [CrossRef] [PubMed]
- Bobrovitz, N.; Ware, H.; Ma, X.; Li, Z.; Hosseini, R.; Cao, C.; Selemon, A.; Whelan, M.; Premji, Z.; Issa, H.; et al. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the omicron variant and severe disease: A systematic review and meta-regression. Lancet Infect. Dis. 2023, 23, 556–567. [Google Scholar] [CrossRef]
- Altarawneh, H.H.; Chemaitelly, H.; Ayoub, H.H.; Tang, P.; Hasan, M.R.; Yassine, H.M.; Al-Khatib, H.A.; Smatti, M.K.; Coyle, P.; Al-Kanaani, Z.; et al. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022, 387, 21–34. [Google Scholar] [CrossRef]
- Crotty, S. Hybrid immunity, COVID-19 vaccine responses provide insights into how the immune system perceives threats. Science 2021, 372, 1392–1393. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.S.; Fukunaga, A.; Yamamoto, S.; Tanaka, A.; Matsuda, K.; Kimura, M.; Kamikawa, A.; Kito, Y.; Maeda, K.; Ueda, G.; et al. SARS-CoV-2 specific T cell and humoral immune responses upon vaccination with BNT162b2: A 9 months longitudinal study. Sci. Rep. 2022, 12, 15447. [Google Scholar] [CrossRef]
- Avis 9721—COVID-19 Vaccination Automne-Hiver Saison 2022–2023. 2022. Available online: https://www.health.belgium.be/fr/node/41634 (accessed on 5 March 2023).
- MacDonald, N.E.J.V. SAGE Working Group on Vaccine Hesitancy. Vaccine hesitancy: Definition, scope and determinants. Vaccine 2015, 33, 4161–4164. [Google Scholar] [CrossRef] [PubMed]
- Covid Vaccinations Belgique. 3 March 2023. Available online: https://covid-vaccinatie.be/fr (accessed on 5 March 2023).
- Lazarus, J.V.; Wyka, K.; White, T.M.; Picchio, C.A.; Gostin, L.O.; Larson, H.J.; Rabin, K.; Ratzan, S.C.; Kamarulzaman, A.; El-Mohandes, A. A survey of COVID-19 vaccine acceptance across 23 countries in 2022. Nat. Med. 2023, 29, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yu, M.; Fu, G.; Lan, G.; Li, L.; Yang, J.; Qiao, Y.; Zhao, J.; Qian, H.-Z.; Zhang, X.; et al. Willingness to Receive COVID-19 Vaccination among People Living with HIV and AIDS in China: Nationwide Cross-sectional Online Survey. JMIR Public Health Surveill. 2021, 7, e31125. [Google Scholar] [CrossRef]
- Lyons, N.; Bhagwandeen, B.; Edwards, J. Factors Affecting COVID-19 Vaccination Intentions among Patients Attending a Large HIV Treatment Clinic in Trinidad Using Constructs of the Health Belief Model. Vaccines 2022, 11, 4. [Google Scholar] [CrossRef]
- Ekstrand, M.L.; Heylen, E.; Gandhi, M.; Steward, W.T.P.; Pereira, M.M.; Srinivasan, K. COVID-19 vaccine hesitancy among PLWH in South India: Implications for Vaccination Campaigns. J. Acquir. Immune Defic. Syndr. 2021, 88, 421–425. [Google Scholar] [CrossRef]
- Antinori, A.; Cicalini, S.; Meschi, S.; Bordoni, V.; Lorenzini, P.; Vergori, A.; Lanini, S.; De Pascale, L.; Matusali, G.; Mariotti, D.; et al. Humoral and cellular immune response elicited by mRNA vaccination against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in people living with human COVID-19 immunogenicity and safety in people with HIV Costiniuk et al. F9 immunodeficiency virus receiving antiretroviral therapy based on current CD4 T-lymphocyte count. Clin. Infect. Dis. 2022, 75, e552–e563. [Google Scholar] [PubMed]
- Gianserra, L.; Dona, M.G.; Giuliani, E.; Stingone, C.; Pontone, M.; Buonomini, A.R.; Giuliani, M.; Pimpinelli, F.; Morrone, A.; Latini, A. Immunogenicity and Safety of BNT162b2 Homologous Booster Vaccination in People Living with HIV under Effective cART. Vaccines 2022, 10, 1243. [Google Scholar] [CrossRef]
- Lapointe, H.R.; Mwimanzi, F.; Cheung, P.K.; Stingone, C.; Pontone, M.; Buonomini, A.R.; Giuliani, M.; Pimpinelli, F.; Morrone, A.; Latini, A.; et al. Antibody response durability following three-dose COVID-19 vaccination in people with HIV receiving suppressive ART. AIDS 2023, 37, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Vergori, A.; Cozzi Lepri, A.; Cicalini, S.; Matusali, G.; Bordoni, V.; Lanini, S.; Meschi, S.; Iannazzo, R.; Mazzotta, V.; Colavita, F.; et al. Immunogenicity to COVID-19 mRNA vaccine third dose in people living with HIV. Nat. Commun. 2022, 13, 4922. [Google Scholar] [CrossRef] [PubMed]
- Aguolu, O.G.; Malik, A.A.; Ahmed, N.; Omer, S.B. Overcoming Vaccine Hesitancy for Future COVID-19 and HIV Vaccines: Lessons from Measles and HPV Vaccines. Curr. HIV/AIDS Rep. 2022, 19, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Mullender, C.; da Costa, K.A.S.; Alrubayyi, A.; Pett, S.L.; Peppa, D. SARS-CoV-2 immunity and vaccine strategies in people with HIV. Oxf. Open Immunol. 2022, 3, iqac005. [Google Scholar] [CrossRef] [PubMed]

| Variable | All (n = 131) | PLWH (n = 80) | HCWs (n = 51) | p-Value |
|---|---|---|---|---|
| Gender | ||||
| Male | 54 (41.2) | 43 (53.8) | 11 (21.6) | 0.0003 |
| Age (Years) | 44.6 ± 10.5 | 45.6 ± 10.7 | 43.0 ± 10.0 | 0.18 |
| 18–29 | 6 (4.6) | 4 (5.0) | 2 (3.9) | |
| 30–39 | 46 (35.1) | 24 (30.0) | 22 (43.1) | |
| 40–49 | 34 (26.0) | 21 (26.2) | 13 (25.5) | |
| 50–59 | 32 (24.4) | 22 (27.5) | 10 (19.6) | |
| ≥60 | 13 (9.9) | 9 (11.3) | 4 (7.8) | |
| BMI (kg/m2) | 26.9 ± 6.1, n = 130 | 27.5 ± 5.6 | 25.9 ± 6.9, n = 50 | 0.13 |
| Underweight (<18.5) | 2 (1.5) | 0 (0.0) | 2 (4.0) | |
| Normal range (18.5–24.9) | 51 (39.2) | 29 (36.2) | 22 (44.0) | |
| Overweight (25–29.9) | 51 (39.2) | 34 (42.5) | 17 (34.0) | |
| Obese (≥30) | 26 (20.0) | 17 (21.3) | 9 (18.0) | |
| Ethnicity | - | |||
| Caucasian | - | 34 (42.5) | - | |
| African | - | 41 (51.3) | - | |
| Other | - | 5 (6.2) | - | |
| Medical history | ||||
| Diabetes mellitus | 6 (4.6) | 5 (6.2) | 1 (2.0) | 0.40 |
| Hypertension | 25 (19.1) | 18 (22.5) | 7 (13.7) | 0.21 |
| Heart failure coronary artery disease | 2 (1.5) | 2 (2.5) | 0 (0.0) | - |
| Stroke | 1 (0.8) | 1 (1.2) | 0 (0.0) | - |
| Liver disease | 1 (0.8) | 1 (1.2) | 0 (0.0) | - |
| Kidney disease | 0 (0.0) | 0 (0.0) | 0 (0.0) | - |
| Chronic lung disease | 1 (0.8) | 1 (1.2) | 0 (0.0) | - |
| Asthma | 3 (2.3) | 0 (0.0) | 3 (5.9) | 0.0028 |
| Autoimmune disease | 2 (1.5) | 0 (0.0) | 2 (3.9) | - |
| Hematological cancer | 1 (0.8) | 0 (0.0) | 1 (2.0) | - |
| Non hematological cancer | 11 (8.4) | 7 (8.8) | 4 (7.8) | 1.0 |
| Solid-organ/cell transplantation | 0 (0.0) | 0 (0.0) | 0 (0.0) | - |
| Immunosuppressive drugs | - | |||
| Corticosteroids | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Other | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Previous SARS-CoV-2 infection (before T0) | ||||
| Questionnaire | 29 (22.1) | 14 (17.5) | 15 (29.4) | 0.11 |
| Positive anti-N antibody | 40 (30.8), n = 131 | 30 (37.5) | 10 (20.0), n = 50 | 0.035 |
| SARS-CoV-2 experienced * | 48 (36.6) | 32 (40.0) | 16 (31.4) | 0.32 |
| Previous SARS-CoV-2 infection (before T1) | ||||
| Questionnaire | 33 (25.2) | 15 (18.8) | 18 (35.3) | 0.033 |
| Positive anti-N antibody | 57 (43.9), n = 130 | 40 (50.0) | 17 (34.0), n = 50 | 0.074 |
| SARS-CoV-2 experienced * | 63 (48.1) | 41 (51.2) | 22 (43.1) | 0.37 |
| Experienced (between T0 and T1) | 15 (11.4) | 9 (11.2) | 6 (11.7) | - |
| First vaccine dose | - | |||
| BNT162b2 mRNA (Pfizer) | 120 (91.6) | 69 (86.2) | 51 (100.0) | |
| mRNA-1273 (Moderna) | 4 (3.0) | 4 (5.0) | 0 (0.0) | |
| ChAdOx1-S (Astra Zeneca) | 7 (5.4) | 7 (8.8) | 0 (0.0) | |
| Second vaccine dose | - | |||
| BNT162b2 mRNA (Pfizer) | 120 (91.6) | 69 (86.2) | 51 (100.0) | |
| mRNA-1273 (Moderna) | 4 (3.0) | 4 (5.0) | 0 (0.0) | |
| ChAdOx1-S (Astra Zeneca) | 7 (5.4) | 7 (8.8) | 0 (0.0) | |
| Third vaccine dose | - | |||
| BNT162b2 mRNA (Pfizer) | 93 (71.0) | 42 (52.5) | 51 (100.0) | |
| mRNA-1273 (Moderna) | 38 (29.0) | 38 (47.5) | 0 (0.0) | |
| Time between first and second vaccine dose (weeks) | 4.0 (3.0–5.0) | 5.0 (4.4–5.0) | 3.0 (3.0–3.1) | <0.0001 |
| Time between second vaccine dose and sample at T0 (weeks) | 24 (24–26) | 25 (23–27) | 24 (24–24) | 0.014 |
| Time between second and third vaccine dose (weeks) | 32 (26–38) | 27 (25-31) | 38 (35–39) | <0.0001 |
| Time between third vaccine dose and sample at T1 (weeks) | 3.7 (2.9–4.7) | 2.4 (3.1–3.9) | 4.7 (4.0–8.0) | <0.0001 |
| Time between T0 and T1 (weeks) | 7 (4–19) | 5 (4–6) | 19 (18–19) | <0.0001 |
| HIV infection | - | |||
| HIV-1 | - | 79 (98.8) | - | |
| HIV-2 | - | 1 (1.2) | - | |
| Time at T0 since HIV diagnosis (years) | 11 (6.5–18) | - | ||
| <1 | - | 1 (1.2) | - | |
| 1–5 | - | 17 (21.3) | - | |
| 6–10 | - | 17 (21.3) | - | |
| >10 | - | 45 (56.2) | - | |
| Nadir CD4+ T cell count per μL | - | 292 (166–502) | - | |
| <200 | - | 25 (31.2) | - | |
| ≥200 | - | 55 (68.8) | - | |
| Last CD4+ T cell count per μL (2021 or 2022) | 743 (592–940) | - | ||
| <350 | - | 3 (3.7) | - | |
| 350–499 | - | 11 (13.8) | - | |
| ≥500 | - | 66 (82.5) | - | |
| CD4/CD8 ratio, n = 117 | 1.1 ± 0.57 | |||
| 0.6–1 | - | 26 (32.5) | - | |
| >1 | - | 38 (47.5) | - | |
| Last plasma viral load copies/mL | <20 (<20–<20) | - | ||
| <50 | - | 75 (93.8) | - | |
| ≥50 | - | 5 (6.2) | - | |
| ART regimen | ||||
| Dual therapy | - | 26 (32.5) | - | |
| NRTI + INI | - | 22 (27.5) | - | |
| NNRTI + INI | - | 4 (5.0) | - | |
| >2 ART | - | 54 (67.5) | - | |
| NRTI + NRTI + INI | - | 35 (43.8) | - | |
| NRTI + NRTI + NNRTI | - | 13 (16.2) | - | |
| NRTI + NRTI + PI | - | 2 (2.5) | - | |
| NRTI + NRTI + PI + INI | - | 2 (2.5) | - | |
| NRTI + INI + PI | - | 1 (1.2) | - | |
| MVC + NRTI + INI + PI | - | 1 (1.2) | - | |
| Time on ART (years) | - | 10.7 ± 6.9 | - | |
| CMV IgG positive | - | 70 (97.2), n = 72 | - | |
| HBsAg positive | - | 1 (1.2) | - | |
| HCV-Ab positive | - | 3 (3.8) | - | |
| Influenza vaccine the same year | - | 33 (25.2) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Moussaoui, M.; Desmecht, S.; Lambert, N.; Maes, N.; Braghini, J.; Marechal, N.; Quintana, C.; Briquet, K.; Gofflot, S.; Toussaint, F.; et al. Cluster Analysis Identifies Distinct Patterns of T-Cell and Humoral Immune Responses Evolution Following a Third Dose of SARS-CoV-2 Vaccine in People Living with HIV. Viruses 2023, 15, 1435. https://doi.org/10.3390/v15071435
El Moussaoui M, Desmecht S, Lambert N, Maes N, Braghini J, Marechal N, Quintana C, Briquet K, Gofflot S, Toussaint F, et al. Cluster Analysis Identifies Distinct Patterns of T-Cell and Humoral Immune Responses Evolution Following a Third Dose of SARS-CoV-2 Vaccine in People Living with HIV. Viruses. 2023; 15(7):1435. https://doi.org/10.3390/v15071435
Chicago/Turabian StyleEl Moussaoui, Majdouline, Salomé Desmecht, Nicolas Lambert, Nathalie Maes, Joachim Braghini, Nicole Marechal, Céline Quintana, Karine Briquet, Stéphanie Gofflot, Françoise Toussaint, and et al. 2023. "Cluster Analysis Identifies Distinct Patterns of T-Cell and Humoral Immune Responses Evolution Following a Third Dose of SARS-CoV-2 Vaccine in People Living with HIV" Viruses 15, no. 7: 1435. https://doi.org/10.3390/v15071435





