Forecasting Outbreaks of Hantaviral Disease: Future Directions in Geospatial Modeling
Abstract
:1. Introduction
2. Forecasting Outbreaks
3. Orthohantavirus Host Specificity
4. Fitness Effects
5. Orthohantavirus Modes of Transmission
6. Host/Human Interactions
7. Environmental Correlates of Risk
8. Forecasting Orthohantavirus Risk
9. Host/Virus Sampling Design
10. Future Directions
- Serology such as IgG ELISA should be able to demonstrate a correspondence between the infection status and the threshold titer used to identify hosts that have been infected. Serology (if accurate) represents prevalent infection data rather than incident infection. Longitudinal studies are best able to identify incidence rates. Incident data is the measure of current virus circulation critical to outbreaks.
- Virus recovery from hosts is the gold standard to establish if a species is a host. Full length sequences of Orthohantaviruses are critical in this regard [28]. To the extent it is practical, virus should also recovered. Establishing a vertebrate species as a host requires evidence for population level rates of infection so that one can distinguish a host species from an incidental spillover from a nearby host [12]. Viral sequencing among different host species is important to establish whether multiple vertebrates serve as sources for a single virus and would need to be monitored as potential sources of human infection.
- Host ecology studies need to expand beyond local population surveys to incorporate metapopulation structure of natural populations. This requires data on local birth-death rates as well as immigration-emigration data [53,55]. Differentiating death from dispersal will be key in understanding fitness effects on subpopulations of hosts most likely to disperse virus.
- Study design of survey methods that introduce biases need to be evaluated. Nearly every study can provide some important clue about virus persistence and distribution, but some designs are not appropriate for the conclusions that need to be reached.
- Ensemble SDMs are among the most efficient ways to extend local knowledge about viral levels in host populations to nearby humans. Ways to address biased sampling impacts on SDMs have received great attention but need to be better integrated into practical applications of vector borne and zoonotic disease forecasting.
- Future developments of machine learning and AI represent the best way forward to make forecasting practical in public health of these agents. It will be critical for developers to intimately understand when machine learning approaches improve analyses and whether AI is robust to deviations from the conditions for which the models were developed, and therefore if they will be of value.
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, H.W.; Lee, P.W.; Johnson, K.M. Isolation of etiologic agent of Korean hemorrhagic-fever. J. Infect. Dis. 1978, 137, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Baek, L.J.; Johnson, K.M. Isolation of Hantaan virus, the etiologic agent of Korean hemorrhagic-fever, from wild urban rats. J. Infect. Dis. 1982, 146, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Schmaljohn, C.S.; Dalrymple, J.M. Analysis of Hantaan virus RNA: Evidence for a new genus of Bunyaviridae. Virology 1983, 131, 482–491. [Google Scholar] [CrossRef]
- Schmaljohn, C.S.; Hasty, S.E.; Harrison, S.A.; Dalrymple, J.M. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 1983, 148, 1005–1012. [Google Scholar] [CrossRef]
- Schmaljohn, C.S.; Hasty, S.E.; Dalrymple, J.M.; LeDuc, J.W.; Lee, H.W.; von Bonsdorff, C.-H.; Brummer-Korvenkontio, M.; Vaheri, A.; Tsai, T.F.; Regnery, H.L.; et al. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science 1985, 227, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Gajdusek, D.C. Virus hemorrhagic fevers. Special reference to hemorrhagic fever with renal syndrome (epidemic hemorrhagic fever). J. Pediatr. 1962, 60, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Khabbaz, R.F.; Armstrong, L.R.; Holman, R.C.; Bauer, S.P.; Graber, J.; Strine, T.; Miller, G.; Reef, S.; Tappero, J.; et al. Hantavirus pulmonary syndrome: The first 100 US cases. J. Infect. Dis. 1996, 173, 1297–1303. [Google Scholar] [CrossRef]
- Yates, T.; Mills, J.N.; Parmenter, C.A.; Ksiazek, T.G. The ecology and evolutionary history of an emergent disease: Hantavirus pulmonary syndrome. Bioscience 2002, 52, 989–998. [Google Scholar] [CrossRef] [Green Version]
- Yanagihara, R.; Gu, S.H.; Arai, S.; Kang, H.J.; Song, J.-W. Hantaviruses: Rediscovery and new beginnings. Virus Res. 2014, 187, 6–14. [Google Scholar] [CrossRef] [Green Version]
- Mull, N.; Jackson, R.; Sironen, T.; Forbes, K.M. Ecology of neglected rodent-borne American Orthohantaviruses. Pathogens 2020, 9, 325. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Reported Cases of Hantavirus Disease. Available online: https://www.cdc.gov/hantavirus/surveillance/index.html (accessed on 25 May 2023).
- Childs, J.E.; Ksiazek, T.G.; Spiropoulou, C.F.; Krebs, J.W.; Morzunov, S.; Maupin, G.O.; Gage, K.L.; Rollin, P.E.; Sarisky, J.; Enscore, R.E.; et al. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the Southwestern United States. J. Infect. Dis. 1994, 169, 1271–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmenter, R.R.; Glass, G.E. Hantavirus outbreaks in the American Southwest: Propagation and retraction of rodent and virus diffusion waves from sky-island refugia. Int. J. Mod. Phys. B 2022, 36, 2140052. [Google Scholar] [CrossRef]
- Morin, C.W.; Semenza, J.C.; Trtanj, J.M.; Glass, G.E.; Boyer, C.; Ebi, K.L. Unexplored opportunities: Use of climate-and weather-driven early warning systems to reduce the burden of infectious diseases. Curr. Envir. Health Rep. 2018, 5, 430–438. [Google Scholar] [CrossRef] [PubMed]
- LeDuc, J.W. The power of we. Viruses 2023, 15, 921. [Google Scholar] [CrossRef]
- Murray, K.O.; Mertens, E.; Despres, P. West Nile virus and its emergence in the United States of America. Vet. Res. 2010, 41, 67. [Google Scholar] [CrossRef] [Green Version]
- Lazear, H.M.; Diamond, M.S. Zika virus: New clinical syndromes and its emergence in the Western hemisphere. J. Virol. 2016, 90, 10. [Google Scholar] [CrossRef] [Green Version]
- Grais, R.F.; Ellis, J.H.; Glass, G.E. Assessing the impact of airline travel on the geographic spread of pandemic influenza. European J. Epidemiol. 2003, 18, 1065–1072. [Google Scholar] [CrossRef]
- National Weather Service, Climate Prediction Center. Climate Prediction Center—Seasonal Outlook. Available online: noaa.gov (accessed on 25 May 2023).
- Famine Early Warning System. Available online: https://fews.net/ (accessed on 25 May 2023).
- California Mosquito-Borne Virus Surveillance & Response Plan. California Department of Health Mosquito & Vector Control Association of California University of California. Available online: https://westnile.ca.gov/ (accessed on 25 May 2023).
- Korch, G.W.; Childs, J.E.; Glass, G.E.; LeDuc, J.W. Spatial and temporal analyses and host range of hantaviral infections within small mammal communities of Baltimore, Maryland USA. Am. J. Trop. Med. Hyg. 1989, 41, 230–240. [Google Scholar] [CrossRef]
- Childs, J.E.; Glass, G.E.; Korch, G.W.; LeDuc, J.W. The ecology and epizootiology of hantaviral infections in small mammal communities of Baltimore: A review and synthesis. Bull. Soc. Vector Ecol. 1988, 13, 1–9. [Google Scholar]
- Arthur, R.R.; Lofts, R.; Gomez, J.; Glass, G.E.; LeDuc, J.W.; Childs, J.E. Grouping of hantaviruses by S genome segment PCR and amplification of viral RNA from wild-caught rats. Am. J. Trop. Med. Hyg. 1992, 46, 410–424. [Google Scholar]
- Song, J.W.; Kang, H.J.; Song, K.J.; Truong, T.T.; Benneft, S.N.; Arai, S.; Truong, N.U.; Yanagihara, R. Newfound hantavirus in Chinese mole hantavirus in Chinese mole shrew, Vietnam. Emerg. Infect. Dis. 2007, 13, 1784–1787. [Google Scholar] [CrossRef]
- Guo, W.P.; Lin, X.D.; Wang, W.; Tian, J.H.; Cong, M.L.; Zhang, H.L.; Wang, M.R.; Zhou, R.H.; Wang, J.B.; Li, M.H.; et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores and rodents. PLoS Pathog. 2013, 9, e1003159. [Google Scholar] [CrossRef] [Green Version]
- Mang, S.; Xian-Dan, L.; Xiao, C.; Jun-Hua, T.; Liang-Jun, C.; Kun, L.; Wen, W.; John-Sebastian, E.; Jin-Jin, S.; Li, L.; et al. Evolutionary history of vertebrate RNA viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Laenen, L.; Vergote, V.; Calsiher, C.H.; Klempa, B.; Klingstrom, J.; Kuhn, J.H.; Maes, P. Hantaviridae: Current classification and future perspectives. Viruses 2019, 11, 788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childs, J.E.; Korch, G.W.; Glass, G.E.; Shah, K.V.; LeDuc, J.W. Epizootiology of Hantavirus infections in Baltimore: Isolation of a virus from wild rats, and characteristics of infected rat populations. Am. J. Epidemiol. 1987, 126, 55–68. [Google Scholar] [CrossRef]
- Klempa, B. Reassortment events in the evolution of hantaviruses. Virus Genes 2018, 54, 638–646. [Google Scholar] [CrossRef] [Green Version]
- Turner, R.W.; Padmowirjono, S.; Martoprawiro, S. Dynamics of the plague transmission cycle in Central Java. (Ecology of the mammalian hosts with special reference to Rattus exulans). Bull. Health Stud. Indones. 1975, 3, 41–71. [Google Scholar]
- Allen, L.J.S.; Wesley, C.L.; Owen, R.D.; Goodin, D.G.; Koch, D.; Jonsson, C.B.; Chu, Y.-K.; Paige, R.L. A habitat-based model for the spread of hantavirus between reservoir and spillover species. J. Theor. Biol. 2009, 260, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Childs, J.E.; Krebs, J.W.; Ksiazek, T.G.; Maupin, G.O.; Gage, K.L.; Rollin, P.E.; Zeitz, P.S.; Sarisky, J.; Enscore, R.E.; Butler, J.C.; et al. A household-based, case-control study of environmental factors associated with hantavirus pulmonary syndrome in the Southwestern United States. Am. J. Trop. Med. Hyg. 1995, 52, 393–397. [Google Scholar] [CrossRef]
- Allen, L.J.; Brown, V.L.; Jonsson, C.B.; Klein, S.K.; Laverty, S.M.; Magwedere, K.; Owen, J.C.; Van den Driessche, P. Mathematical modeling of viral zoonoses in wildlife. Nat. Resour. Model. 2012, 25, 5–51. [Google Scholar] [CrossRef]
- Mills, J.N.; Ksiazek, T.G.; Peters, C.J.; Childs, J.E. Long-term studies of hantavirus reservoir populations in the Southwestern United States: A synthesis. Emerg. Infect. Dis. 1999, 5, 135–142. [Google Scholar] [CrossRef]
- Childs, J.E.; Glass, G.E.; Korch, G.W.; LeDuc, J.W. Effects of hantaviral infection on survival, growth and fertility in wild rat (Rattus norvegicus) populations of Baltimore, Maryland. J. Wldlf Dis. 1989, 25, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Luis, A.D.; Douglass, R.J.; Hudson, P.J.; Mills, J.N.; Bjornstad, O.N. Sin Nombre hantavirus decreases survival of male deer mice. Oecologia 2012, 169, 431–439. [Google Scholar] [CrossRef]
- Tuttle, M.D. Population ecology of the gray bat (Myotis grisescens): Factors influencing growth and survival of newly volant young. Ecology 1976, 57, 587–595. [Google Scholar] [CrossRef]
- Eby, P. Seasonal movements of grey-headed flying-foxes Pteropus poliocephalus (Chiropter: Pteropodidae), from two maternity camps in northern New South Wales. Wildlf. Res. 1991, 18, 547–559. [Google Scholar] [CrossRef]
- Armien, B.; Ortiz, P.L.; Gonzalez, P.; Cumbrera, A.; Rivero, A.; Avila, M.; Armien, A.G.; Koster, F.; Glass, G. Spatial-temporal distribution of Hantavirus rodent-borne infection by Oligoryzomys fulvescens in the Agua Buena region—Panama. PLoS NTD 2016, 10, e0004460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juan, E.; Levis, S.; Pini, N.; Polop, J.; Steinmann, A.R.; Provensal, M.C. Mechanisms of hantavirus transmission in Oligoryzomys longicaudatus. Ecohealth 2019, 16, 671–681. [Google Scholar] [CrossRef]
- Clement, J.; Le Duc, J.W.; Lloyd, G.; Reynes, J.-M.; McElhinney, L.; Van Ranst, M.; Lee, H.-W. Wild rats, laboratory rats, pet rats: Global Seoul hantavirus disease revisited. Viruses 2019, 11, 652. [Google Scholar] [CrossRef] [Green Version]
- Traub, R.; Wisseman, C.L., Jr. Korean hemorrhagic fever. J. Infect. Dis. 1978, 138, 267–272. [Google Scholar] [CrossRef]
- Lee, H.W.; Lee, P.W.; Baek, L.J.; Song, C.K.; Seong, I.W. Intraspecific transmission of Hantaan Virus, etiologic agent of Korean Hemorrhagic Fever, in the rodent Apodemus agrarius. Am. J. Trop. Med. Hyg. 1981, 30, 1106–1112. [Google Scholar] [CrossRef]
- Lee, H.W.; Johnson, K.M. Laboratory-acquired infections with Hantaan virus, the etiologic agent of Korean hemorrhagic-fever. J. Infect. Dis. 1982, 146, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, J.; Yamanouchi, T.; Dohmae, K.; Miyamoto, H.; Takahaski, M.; Yamanishi, K.; Kurata, T.; Lee, H.W. Control of laboratory acquired hemorrhagic fever with renal syndrome (HFRS) in Japan. Lab. Anim. Sci. 1987, 37, 431–436. [Google Scholar]
- Desmyter, J.; LeDuc, J.W.; Johnson, K.M.; Brasseur, F.; Deckers, C.; van Ypersele, S.C. Laboratory rat associated outbreak of haemorrhagic fever with renal syndrome due to Hantaan-like virus in Belgium. Lancet 1983, 8365–8366, 1445–1448. [Google Scholar] [CrossRef]
- Klein, S.L.; Bird, B.H.; Glass, G.E. Sex differences in Seoul virus infection are not related to adult sex steroid concentrations in Norway rats. J. Virol. 2000, 74, 8213–8217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, G.R.; McKee, K.T., Jr. Pathogenesis of Hantaan Virus infection in suckling mice: Clinical, virologic, and serologic observations. Am. J. Trop Med. Hyg. 1985, 34, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, R.; Amyx, H.L.; Gajdusek, D.C. Experimental infection with Puumala virus, the etiologic agent of nephropathia epidemica, in bank voles (Clethrionomys glareolus). J. Virol. 1985, 55, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-K.; Takashima, I.; Hashimoto, N. Role of maternal antibody in protection from hemorrhagic fever with renal syndrome virus infection in rats. Arch. Virol. 1988, 103, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Bascompte, J. Extinction thresholds: Insights from simple models. Ann. Zool. Fennici. 2003, 40, 99–114. Available online: https://www.jstor.org/stable/23736518 (accessed on 25 May 2023).
- Mills, J.N.; Ksiazek, T.G.; Ellis, B.A.; Rollin, P.E.; Nichol, S.T.; Yates, T.L.; Gannon, W.L.; Levy, C.E.; Engelthaler, D.M.; Davis, T.; et al. Patterns of association with host and habitat: Antibody reactive with Sin Nombre virus in small mammals in the major biotic communities of the Southwestern United States. Am. J. Trop. Med. Hyg. 1997, 56, 273–284. [Google Scholar] [CrossRef]
- Parmenter, C.A.; Yates, T.L.; Parmenter, R.R.; Dunnum, J.L. Statistical sensitivity for detection of spatial and temporal patterns in rodent population densities. Emerg. Inf. Dis. 1999, 5, 118–125. [Google Scholar] [CrossRef]
- Glass, G.E.; Livingstone, W.; Mills, J.N.; Hlady, W.G.; Fine, J.B.; Rollin, P.E.; Ksiazek, T.G.; Peters, C.J.; Childs, J.E. Black Creek Canal virus infection in Sigmodon hispidus in southern Florida. Am. J. Trop. Med. Hyg. 1998, 59, 699–703. [Google Scholar] [CrossRef] [Green Version]
- Hinson, E.; Shone, S.; Zink, M.C.; Glass, G.E.; Klein, S.A. Wounding: The primary mode of Seoul virus transmission among male Norway rats. Am. J. Trop. Med. Hyg. 2004, 70, 310–317. [Google Scholar] [CrossRef]
- Nuzum, E.O.; Rossi, C.A.; Stephenson, E.H.; LeDuc, J.W. Aerosol transmission of Hantaan and related viruses to laboratory rats. Am. J. Trop. Med. Hyg. 1988, 38, 636–640. [Google Scholar] [CrossRef]
- Glass, G.E.; Childs, J.E.; Korch, G.W.; LeDuc, J.W. Association of intraspecific aggression and hantavirus infection in wild rats (Rattus norvegicus). Epidemiol. Infect. 1988, 101, 459–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childs, J.E.; Glass, G.E.; LeDuc, J.W. Rodent sightings and contacts in an inner-city population of Baltimore, Maryland USA. Bull. Soc. Vector Ecol. 1991, 16, 245–255. [Google Scholar]
- Klein, S.L.; Zink, M.L.; Glass, G.E. Seoul virus infection increases aggressive behaviour in male Norway rats. Anim. Behav. 2004, 67, 421–429. [Google Scholar] [CrossRef]
- Abbott, K.D.; Ksiazek, T.G.; Mills, J.N. Long-term hantavirus persistence in rodent populations in central Arizona. Emerg. Inf. Dis. 1999, 5, 102–112. [Google Scholar] [CrossRef]
- Vitek, C.R.; Breiman, R.F.; Ksiazek, T.G.; Rollin, P.E.; McLaughlin, J.C.; Umland, E.T.; Nolte, K.B.; Loera, A.; Sewell, C.M.; Peters, C.J. Evidence against person to person transmission of hantavirus to healthcare workers. Clin. Infect. Dis. 1996, 22, 824–826. [Google Scholar] [CrossRef]
- Childs, J.E. Zoonotic viruses of wildlife: Hither from yon. In Emergence and Control of Zoonotic Viral Encephalitides; Archives of Virology; Springer: Berlin/Heidelberg, Germany, 2004; Volume 18, pp. 1–11. [Google Scholar] [CrossRef]
- Flanagan, M.L.; Parrish, C.R.; Cobey, S.; Glass, G.E.; Bush, R.M.; Leighton, T.J. Anticipating the species jump: Surveillance for emerging viral threats. Zoonoses Publ. Health 2012, 59, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Enria, D.; Padula, P.; Segura, E.L.; Pini, N.; Edelstein, A.; Posse, C.R.; Weissenbacher, M.C. Hantavirus pulmonary syndrome in Argentina. Possibility of person to person transmission. Medicina 1996, 56, 709–711. [Google Scholar] [PubMed]
- Martinez-Valdebenito, C.; Calvo, M.; Vial, C.; Mansilla, R.; Marco, C.; Palma, R.E.; Vial, P.A.; Valdivieso, F.; Mertz, G.; Ferres, M. Person-to-person household and nosocomial transmission of Andes Hantavirus, Southern Chile, 2011. Emerg. Inf. Dis. 2014, 20, 1629–1644. [Google Scholar] [CrossRef] [PubMed]
- Hardestam, J.; Karlsson, M.; Falk, K.I.; Olsson, G.; Klingstrom, J.; Lunkkvist, A. Puumala hantavirus excretion kinetics in Bank voles (Myodes glareolus). Emerg. Inf. Dis. 2008, 14, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.E.; St Jeor, S.C.; Riolo, J.; Otteson, E.W.; Monroe, M.C.; Henderson, W.W.; Ksiazek, T.G.; Rollin, P.E.; Nichol, S.T. Coexistence of several novel hantaviruses in rodents indigenous to North America. Virology 1995, 213, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.J.; Quiroz, E.; Gracia, F.; Sanchez, A.J.; Ksiazek, T.G.; Kitsutani, P.T.; Ruedas, L.A.; Tinnin, D.S.; Caceres, L.; Garcia, A.; et al. Hantavirus pulmonary syndrome in Panama: Identification of novel hantaviruses and their likely reservoirs. Virology 2000, 277, 14–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, R.; Canate, R.; Pascale, J.M.; Dragoo, J.W.; Armien, B.; Armien, A.G.; Koster, F. Confirmation of Choclo virus as the cause of hantavirus cardiopulmonary syndrome and high serum antibody prevalence in Panama. J. Med. Virol. 2010, 82, 1586–1593. [Google Scholar] [CrossRef] [Green Version]
- Knust, B.; Rollin, P.E. Twenty-year summary of surveillance for human hantavirus infections, United States. Emerg. Infect. Dis. 2013, 19, 1934–1937. [Google Scholar] [CrossRef]
- Glass, G.E.; Johnson, J.S.; Hodenbach, G.A.; DiSalvo, C.L.J.; Peters, C.J.; Childs, J.E.; Mills, J.N. Experimental evaluation of rodent exclusion methods to reduce hantavirus transmission to humans in rural housing. Am. J. Trop. Med. Hyg. 1997, 56, 359–364. [Google Scholar] [CrossRef]
- Zeitz, P.S.; Butler, J.C.; Cheek, J.E.S.; Michael, C.; Childs, J.E.; Shands, L.A.; Turner, R.E.; Voorhees, R.E.; Sarisky, J.; Rollin, P.E.; et al. A case-control study of Hantavirus Pulmonary Syndrome during an outbreak in the Southwestern United States. J. Infect. Dis. 1995, 171, 864–870. [Google Scholar] [CrossRef] [Green Version]
- Vittor, A.M.; Armien, B.; Gonzalez, P.; Carrera, J.P.; Dominguez, C.; Valderrama, A.; Glass, G.E.; Beltran, D.; Cisneros, J.; Wang, E.; et al. Epidemiology of emergent Madariaga Encephalitis in a region with endemic Venezuelan Equine Encephalitis: Initial host studies and human cross-sectional study in Darien, Panama. PLoS NTD 2016, 10, e0004554. [Google Scholar] [CrossRef] [Green Version]
- Ostfeld, R.S.; Glass, G.E.; Keesing, F. Landscape epidemiology: An emerging (or re-emerging) discipline. Trends Ecol. Evol. 2005, 20, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Florio, E.N.; Lele, S.R.; Chang, Y.-C.; Sterner, R.; Glass, G.E. Integrating AVHRR satellite data and NOAA ground observations to predict land surface temperature: A statistical approach. Int. J. Remote Sens. 2004, 25, 2979–2994. [Google Scholar] [CrossRef]
- Johansson, M.A.; Glass, G.E. High-resolution spatiotemporal weather models for climate studies. Int. J. Health Geogr. 2008, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Glass, G.E. Health and Disease. In Comprehensive Remote Sensing; Liang, S., Ed.; Elsevier: Oxford, UK, 2018; Volume 9, pp. 268–279. ISBN 9780128032206. [Google Scholar]
- Schaper, T.; Khatami, R.; Southworth, J.; Glass, G.E. Monitoring major crop change trends in agricultural [sic] in Florida. Am. J. GIS. 2022, 11, 23–31. [Google Scholar]
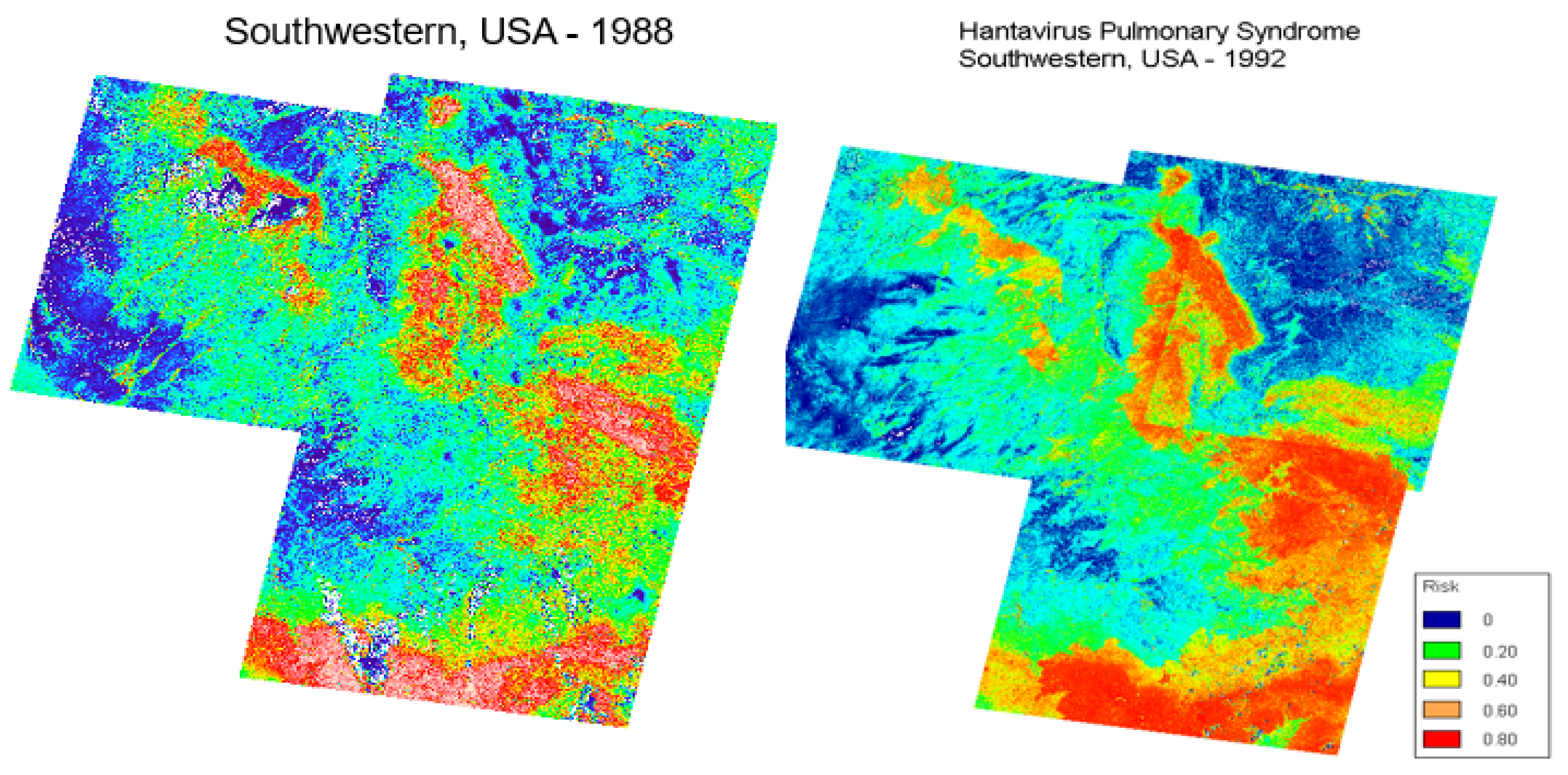
- Glass, G.E.; Cheek, J.E.; Patz, J.A.; Shields, T.M.; Doyle, T.J.; Thoroughman, D.A.; Hunt, D.K.; Enscore, R.E.; Gage, K.L.; Irland, C.; et al. Anticipating risk areas for hantavirus pulmonary syndrome with remotely sensed data: Re-examination of the 1993 outbreak. Emerg. Inf. Dis. 2000, 6, 238–247. [Google Scholar] [CrossRef] [Green Version]
- Glass, G.E.; Shields, T.; Cai, B.; Yates, T.L.; Parmenter, R. Persistently highest risk areas for Hantavirus Pulmonary Syndrome: Potential sites for refugia. Ecol. Appl. 2007, 17, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Glass, G.E.; Shields, T.M.; Parmenter, R.R.; Goade, D.; Mills, J.N.; Cheek, J.; Cook, J.; Yates, T.L. Predicted Hantavirus Risk in 2006 for the Southwestern, U. S.; Museum of Texas Tech University: Lubbock, TX, USA, 2006; pp. 1–14. [Google Scholar] [CrossRef]
- Walsh, A.S.; Louis, T.A.; Glass, G.E. Detecting multiple levels of effect during survey sampling using a Bayesian approach: Point prevalence estimates of hantavirus in cotton rats (Sigmodon hispidus). Ecol. Model. 2007, 205, 29–38. [Google Scholar] [CrossRef]
- Andreo, V.; Glass, G.; Shields, T.; Provensal, C.; Polop, J. Modeling potential distribution of Oligoryzomys longicaudatus, the Andes virus (Genus: Hantavirus) reservoir, in Argentina. EcoHealth 2011, 8, 332–348. [Google Scholar] [CrossRef] [PubMed]
- Glass, G.E.; Yates, T.L.; Fine, J.B.; Shields, T.M.; Kendall, J.B.; Hope, A.G.; Parmenter, C.A.; Peters, C.J.; Ksiazek, T.G.; Li, C.-S.; et al. Satellite imagery characterizes local animal reservoir populations of Sin Nombre virus in Southwestern United States. Proc. Nat. Acad. Sci. USA 2002, 99, 16817–16822. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Ann. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H. Do they? How do they? Why do they differ? On finding reasons for differing performances of species distribution models. Ecography 2009, 32, 66–77. [Google Scholar] [CrossRef]
- Stockwell, D.; Peters, D. The GARP modelling system: Problems and solutions to automated spatial prediction. Int. J. Geogr. Inform. Sci. 1999, 13, 143–158. [Google Scholar] [CrossRef]
- Busby, J.R. BIOCLIM—A bioclimate analysis and prediction system. In Nature Conservation: Cost Effective Biological Surveys and Data Analysis; Margules, C.R., Austin, M.P., Eds.; CSIRO: Canberra, Australia, 1991; pp. 64–68. [Google Scholar]
- Kessler, W.H.; Ganser, C.; Glass, G.E. Modeling the distribution of medically important ticks in Florida. Insects 2019, 10, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, Y.P.; Jarnevich, C.S.; Barnett, D.T.; Monaghan, A.J.; Eisen, R.J. Modeling the present and future geographic distribution of the lone star tick, Amblyomma americanum (Ixodida: Ixodidae), in the Continental United States. Am. J. Trop. Med. Hyg. 2015, 93, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Kessler, W.H.; DeJesus, C.; Wisely, S.M.; Glass, G.E. Ensemble models for tick vectors: Standard surveys compared with convenience samples. Diseases 2022, 10, 32. [Google Scholar] [CrossRef]
- Healy, G.R.; Spielman, A.; Gleason, N. Human Babesiosis: Reservoir of infection on Nantucket Island. Science 1976, 4238, 479–480. [Google Scholar] [CrossRef]
- Glass, G.E. Infectious Disease Ecology [Review]. Bioscience 2009, 59, 263–264. [Google Scholar] [CrossRef]
- Baker, D.J.; Maclean, I.M.D.; Goodall, M.; Gaston, K.J. Correlations between spatial sampling biases and environmental niches affect species distribution models. Global Ecol. Biogeogr. 2022, 31, 1038–1050. [Google Scholar] [CrossRef]
- Zizka, A.; Antonelli, A.; Silvestro, D. Sampbias, a method for quantifying geographic sampling biases in species distribution data. Ecography 2021, 44, 25–32. [Google Scholar] [CrossRef]
- Gutierrez-Velez, V.H.; Wiese, D. Sampling bias mitigation for species occurrence modeling using machine learning methods. Ecol. Inform. 2020, 58, 101091. [Google Scholar] [CrossRef]
- Nelson, K.; Masters Williams, C. Infectious Disease Epidemiology: Theory and Practice, 3rd ed.; Jones and Bartlett: Boston, MA, USA, 2014. [Google Scholar]
- Kelsey, J.L.; Whittemore, A.S.; Evans, A.S.; Thompson, W.D. Methods in Observational Epidemiology, 2nd ed.; Oxford University Press: New York, NY, USA, 1996. [Google Scholar]
- Jackson, E.E.; Janitz, A.E.; Carabin, H. A method to create directed acyclic graphs from cycles of transmission of zoonotic and vector-borne infectious agents. Vector Borne Zoo. Dis. 2023, 23, 129–135. [Google Scholar] [CrossRef]
- Fithian, W.; Elith, J.; Hastie, T.; Keith, D.A. Bias correction in species distribution models: Pooling survey and collection data for multiple species. Methods Ecol. Evol. 2015, 6, 424–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, P.; Salinas, T.P.; Salazar, J.R.; Armien, A.G.; Avila, M.; Broce, E.; Cook, J.A.; Colella, J.P.; Dunnum, J.; Glass, G.; et al. Two decades of wildlife surveillance provide a foundation for understanding zoonotic disease dynamics across space and time: Case study of Choclo orthohantaviruses and their wild reservoir. Viruses, 2023; in press. [Google Scholar]
- Law, R.; Illian, J.; Burslem, D.R.F.P.; Gratzer, G.; Gunatilleke, C.V.S.; Gunatilleke, I.A.U.N. Ecological information from spatial patterns of plants: Insights from point process theory. J. Ecol. 2009, 97, 616–628. [Google Scholar] [CrossRef]
- Mollalo, A.; Glass, G.E.; Sofizadeh, A.; Sadeghian, A.; Israel, G.D.; Rashidi, P. GIS-based ecological modeling of the sandfly Phlebotomus papatasi, the main vector of zoonotic cutaneous Leishmaniasis: A machine learning approach. Acta Trop. 2018, 188, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Taesiri, M.R.; Nguyen, G.; Nguyen, A. Visual correspondence-based explanations improve AI robustness and human-IA team accuracy. In Proceedings of the Thirty-sixth Conference on Neural Information Processing Systems, New Orleans, LA, USA, 28 November 2022–9 December 2022; pp. 427–436. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glass, G.E. Forecasting Outbreaks of Hantaviral Disease: Future Directions in Geospatial Modeling. Viruses 2023, 15, 1461. https://doi.org/10.3390/v15071461
Glass GE. Forecasting Outbreaks of Hantaviral Disease: Future Directions in Geospatial Modeling. Viruses. 2023; 15(7):1461. https://doi.org/10.3390/v15071461
Chicago/Turabian StyleGlass, Gregory E. 2023. "Forecasting Outbreaks of Hantaviral Disease: Future Directions in Geospatial Modeling" Viruses 15, no. 7: 1461. https://doi.org/10.3390/v15071461
APA StyleGlass, G. E. (2023). Forecasting Outbreaks of Hantaviral Disease: Future Directions in Geospatial Modeling. Viruses, 15(7), 1461. https://doi.org/10.3390/v15071461




