Structure and Function of Hoc—A Novel Environment Sensing Device Encoded by T4 and Other Bacteriophages
Abstract
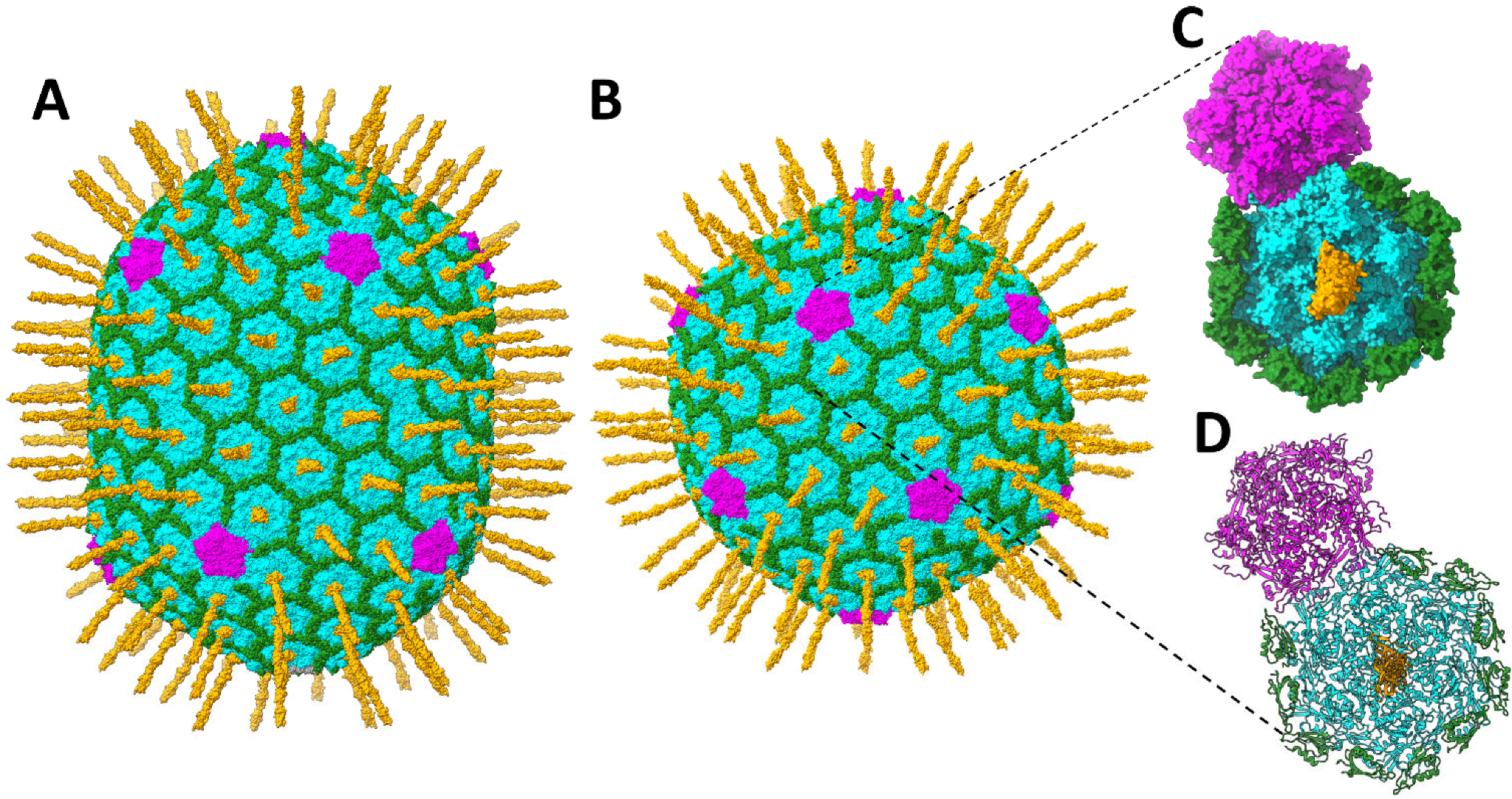
:1. Introduction
2. Materials and Methods
2.1. Hoc Model Building and Refinement
2.2. In Vitro Assembly of Hoc on T4 Phage Capsid
2.3. Plaque Assay to Determine Clustering vs. Dispersion
2.4. Sedimentation Assay to Determine pH Dependent Clustering vs. Dispersion
3. Results
3.1. Model of the T4 Hoc Protein
3.2. Vertex–Proximal Capsomers Showed Preferred Orientations of Bound Hoc
3.3. Structure of the Hoc C-Terminal Domain Attached to the Vertex–Proximal gp23* Hexamers
3.4. Interactions between Hoc C-Terminal Domain and Major Capsid Protein gp23*
3.5. Hoc C-Domain Interactions in Alternative Orientations
3.6. Structural Models of Phage T4 Full-Length Hoc and of Hoc-like Molecules from Other Bacteriophages
3.7. Hoc Facilitates Dispersion of Phage T4 Virion Particles
3.8. Hoc Might Function as an Environmental Sensor
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fokine, A.; Chipman, P.R.; Leiman, P.G.; Mesyanzhinov, V.V.; Rao, V.B.; Rossmann, M.G. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA 2004, 101, 6003–6008. [Google Scholar] [CrossRef]
- Fang, Q.; Tang, W.-C.; Fokine, A.; Mahalingam, M.; Shao, Q.; Rossmann, M.G.; Rao, V.B. Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly. Proc. Natl. Acad. Sci. USA 2022, 119, e2203272119. [Google Scholar] [CrossRef]
- Rao, V.B.; Fokine, A.; Fang, Q.; Shao, Q. Bacteriophage T4 Head: Structure, Assembly, and Genome Packaging. Viruses 2023, 15, 527. [Google Scholar] [CrossRef] [PubMed]
- Black, L.W.; Rao, V.B. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv. Virus Res. 2012, 82, 119–153. [Google Scholar]
- Leiman, P.G.; Arisaka, F.; van Raaij, M.J.; Kostyuchenko, V.A.; Aksyuk, A.A.; Kanamaru, S.; Rossmann, M.G. Morphogenesis of the T4 tail and tail fibers. Virol. J. 2010, 7, 355. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Rüger, W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Sun, L.; Zhang, Z.; Fokine, A.; Padilla-Sanchez, V.; Hanein, D.; Jiang, W.; Rossmann, M.G.; Rao, V.B. Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-Å resolution and its relevance to the assembly of icosahedral viruses. Proc. Natl. Acad. Sci. USA 2017, 114, E8184–E8193. [Google Scholar] [CrossRef]
- Fokine, A.; Leiman, P.G.; Shneider, M.M.; Ahvazi, B.; Boeshans, K.M.; Steven, A.C.; Black, L.W.; Mesyanzhinov, V.V.; Rossmann, M.G. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA 2005, 102, 7163–7168. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, R.A.; Engel, A.; tenHeggeler, B.; van Driel, R. Gene 20 product of bacteriophage T4 its purification and structure. J. Mol. Biol. 1981, 152, 641–662. [Google Scholar] [CrossRef]
- Fang, Q.; Tang, W.C.; Tao, P.; Mahalingam, M.; Fokine, A.; Rossmann, M.G.; Rao, V.B. Structural morphing in a symmetry-mismatched viral vertex. Nat. Commun. 2020, 11, 1713. [Google Scholar] [CrossRef] [Green Version]
- Rao, V.B.; Fokine, A.; Fang, Q. The remarkable viral portal vertex: Structure and a plausible model for mechanism. Curr. Opin. Virol. 2021, 51, 65–73. [Google Scholar] [CrossRef]
- Ishii, T.; Yanagida, M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 1977, 109, 487–514. [Google Scholar] [CrossRef]
- Fokine, A.; Islam, M.Z.; Zhang, Z.; Bowman, V.D.; Rao, V.B.; Rossmann, M.G. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J. Virol. 2011, 85, 8141–8148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathaliyawala, T.; Islam, M.Z.; Li, Q.; Fokine, A.; Rossmann, M.G.; Rao, V.B. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 2010, 77, 444–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, L.; Fokine, A.; O’Donnell, E.; Rao, V.B.; Rossmann, M.G. Structure of the small outer capsid protein, Soc: A clamp for stabilizing capsids of T4-like phages. J. Mol. Biol. 2010, 395, 728–741. [Google Scholar] [CrossRef] [Green Version]
- Black, L.W.; Showe, M.K.; Steven, A.C. Morphogenesis of the T4 head. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 218–258. [Google Scholar]
- Shivachandra, S.B.; Li, Q.; Peachman, K.K.; Matyas, G.R.; Leppla, S.H.; Alving, C.R.; Rao, M.; Rao, V.B. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine 2007, 25, 1225–1235. [Google Scholar] [CrossRef] [Green Version]
- Tao, P.; Zhu, J.; Mahalingam, M.; Batra, H.; Rao, V.B. Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv. Drug Deliv. Rev. 2019, 145, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.B.; Zhu, J. Bacteriophage T4 as a nanovehicle for delivery of genes and therapeutics into human cells. Curr. Opin. Virol. 2022, 55, 101255. [Google Scholar] [CrossRef]
- Tao, P.; Mahalingam, M.; Marasa, B.S.; Zhang, Z.; Chopra, A.K.; Rao, V.B. In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc. Natl. Acad. Sci. USA 2013, 110, 5846–5851. [Google Scholar] [CrossRef]
- Fokine, A.; Bowman, V.D.; Battisti, A.J.; Li, Q.; Chipman, P.R.; Rao, V.B.; Rossmann, M.G. Cryo-electron microscopy study of bacteriophage T4 displaying anthrax toxin proteins. Virology 2007, 367, 422–427. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Batra, H.; Ananthaswamy, N.; Mahalingam, M.; Tao, P.; Wu, X.; Guo, W.; Fokine, A.; Rao, V.B. Design of bacteriophage T4-based artificial viral vectors for human genome remodeling. Nat. Commun. 2023, 14, 2928. [Google Scholar] [CrossRef] [PubMed]
- Fokine, A.; Battisti, A.J.; Kostyuchenko, V.A.; Black, L.W.; Rossmann, M.G. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: The evolution of pentameric vertex proteins in icosahedral viruses. J. Struct. Biol. 2006, 154, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Black, L.W.; Bisher, M.E.; Steven, A.C. Assembly-dependent conformational changes in a viral capsid protein. Calorimetric comparison of successive conformational states of the gp23 surface lattice of bacteriophage T4. J. Mol. Biol. 1985, 183, 353–364. [Google Scholar] [CrossRef]
- Bateman, A.; Eddy, S.R.; Mesyanzhinov, V.V. A member of the immunoglobulin superfamily in bacteriophage T4. Virus Genes 1997, 14, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, K.; Świtała-Jeleń, K.; Opolski, A.; Górski, A. Possible association between phages, Hoc protein, and the immune system. Arch. Virol. 2006, 151, 209–215. [Google Scholar] [CrossRef]
- Chin, W.H.; Kett, C.; Cooper, O.; Müseler, D.; Zhang, Y.; Bamert, R.S.; Patwa, R.; Woods, L.C.; Devendran, C.; Korneev, D.; et al. Bacteriophages evolve enhanced persistence to a mucosal surface. Proc. Natl. Acad. Sci. USA 2022, 119, e2116197119. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776. [Google Scholar] [CrossRef]
- Fraser, J.S.; Yu, Z.; Maxwell, K.L.; Davidson, A.R. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J. Mol. Biol. 2006, 359, 496–507. [Google Scholar] [CrossRef]
- Dabrowska, K.; Zembala, M.; Boratynski, J.; Switala-Jelen, K.; Wietrzyk, J.; Opolski, A.; Szczaurska, K.; Kujawa, M.; Godlewska, J.; Gorski, A. Hoc protein regulates the biological effects of T4 phage in mammals. Arch. Microbiol. 2007, 187, 489–498. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Sam-Soon, N.; Kassegne, S.; Peters, G.; Bonilla, N.; Hatay, M.; Mourtada, S.; Bailey, B.; Youle, M.; et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 2015, 112, 13675–13680. [Google Scholar] [CrossRef]
- Vernhes, E.; Renouard, M.; Gilquin, B.; Cuniasse, P.; Durand, D.; England, P.; Hoos, S.; Huet, A.; Conway, J.F.; Glukhov, A.; et al. High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid. Sci. Rep. 2017, 7, 41662. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 4, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonine, P.V.; Poon, B.K.; Read, R.J.; Sobolev, O.V.; Terwilliger, T.C.; Urzhumtsev, A.; Adams, P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018, 74 Pt 6, 531–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75 Pt 10, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.; Brookes, D.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [Green Version]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Holm, L. Dali server: Structural unification of protein families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Atkison, C.; Moreland, R.; Liu, M.; Ramsey, J.; Leavitt, J. Complete genome sequence of Serratia phage Muldoon. Microbiol. Resour. Announc. 2020, 9, e01418-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, D.F.; Pye, G.; Bramley, R.; Clark, A.G.; Dyson, T.J.; Hardcastle, J.D. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 1988, 29, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Szermer-Olearnik, B.; Drab, M.; Mąkosa, M.; Zembala, M.; Barbasz, J.; Dąbrowska, K.; Boratyński, J. Aggregation/dispersion transitions of T4 phage triggered by environmental ion availability. J. Nanobiotechnol. 2017, 15, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conley, M.P.; Wood, W.B. Bacteriophage T4 whiskers: A rudimentary environment-sensing device. Proc. Natl. Acad. Sci. USA 1975, 72, 3701–3705. [Google Scholar] [CrossRef]
- Islam, M.Z.; Fokine, A.; Mahalingam, M.; Zhang, Z.; Garcia-Doval, C.; van Raaij, M.J.; Rossmann, M.G.; Rao, V.B. Molecular anatomy of the receptor binding module of a bacteriophage long tail fiber. PLoS Pathog. 2019, 15, e1008193. [Google Scholar] [CrossRef] [Green Version]
- Brümmendorf, T.; Rathjen, F.G. Cell adhesion molecules 1: Immunoglobulin superfamily. Protein Profile 1995, 2, 963–1108. [Google Scholar]
- Aricescu, A.R.; Jones, E.Y. Immunoglobulin superfamily cell adhesion molecules: Zippers and signals. Curr. Opin. Cell Biol. 2007, 19, 543–550. [Google Scholar] [CrossRef]
- Halaby, D.M.; Mornon, J.P. The immunoglobulin superfamily: An insight on its tissular, species, and functional diversity. J. Mol. Evol. 1998, 46, 389–400. [Google Scholar] [CrossRef]
- Fraser, J.S.; Maxwell, K.L.; Davidson, A.R. Immunoglobulin-like domains on bacteriophage: Weapons of modest damage? Curr. Opin. Microbiol. 2007, 10, 382–387. [Google Scholar] [CrossRef]








| Hoc-gp23 in Isometric Capsid | Hoc-gp23 in Prolate Capsid | |
|---|---|---|
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.007 |
| Bond angles (°) | 0.62 | 0.69 |
| Dihedral angles (°) | 4.84 | 13.50 |
| Ramachandran plot | ||
| Favored (%) | 91.1 | 90.3 |
| Allowed (%) | 8.5 | 9.1 |
| Disallowed (%) | 0.4 | 0.6 |
| Rotamers outliers (%) | 0.67 | 5.53 |
| Clashscore | 13.86 | 12.84 |
| CCmask | 0.85 | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fokine, A.; Islam, M.Z.; Fang, Q.; Chen, Z.; Sun, L.; Rao, V.B. Structure and Function of Hoc—A Novel Environment Sensing Device Encoded by T4 and Other Bacteriophages. Viruses 2023, 15, 1517. https://doi.org/10.3390/v15071517
Fokine A, Islam MZ, Fang Q, Chen Z, Sun L, Rao VB. Structure and Function of Hoc—A Novel Environment Sensing Device Encoded by T4 and Other Bacteriophages. Viruses. 2023; 15(7):1517. https://doi.org/10.3390/v15071517
Chicago/Turabian StyleFokine, Andrei, Mohammad Zahidul Islam, Qianglin Fang, Zhenguo Chen, Lei Sun, and Venigalla B. Rao. 2023. "Structure and Function of Hoc—A Novel Environment Sensing Device Encoded by T4 and Other Bacteriophages" Viruses 15, no. 7: 1517. https://doi.org/10.3390/v15071517
APA StyleFokine, A., Islam, M. Z., Fang, Q., Chen, Z., Sun, L., & Rao, V. B. (2023). Structure and Function of Hoc—A Novel Environment Sensing Device Encoded by T4 and Other Bacteriophages. Viruses, 15(7), 1517. https://doi.org/10.3390/v15071517






