Hepatitis Delta Virus–Host Protein Interactions: From Entry to Egress
Abstract
:1. Introduction
2. Review
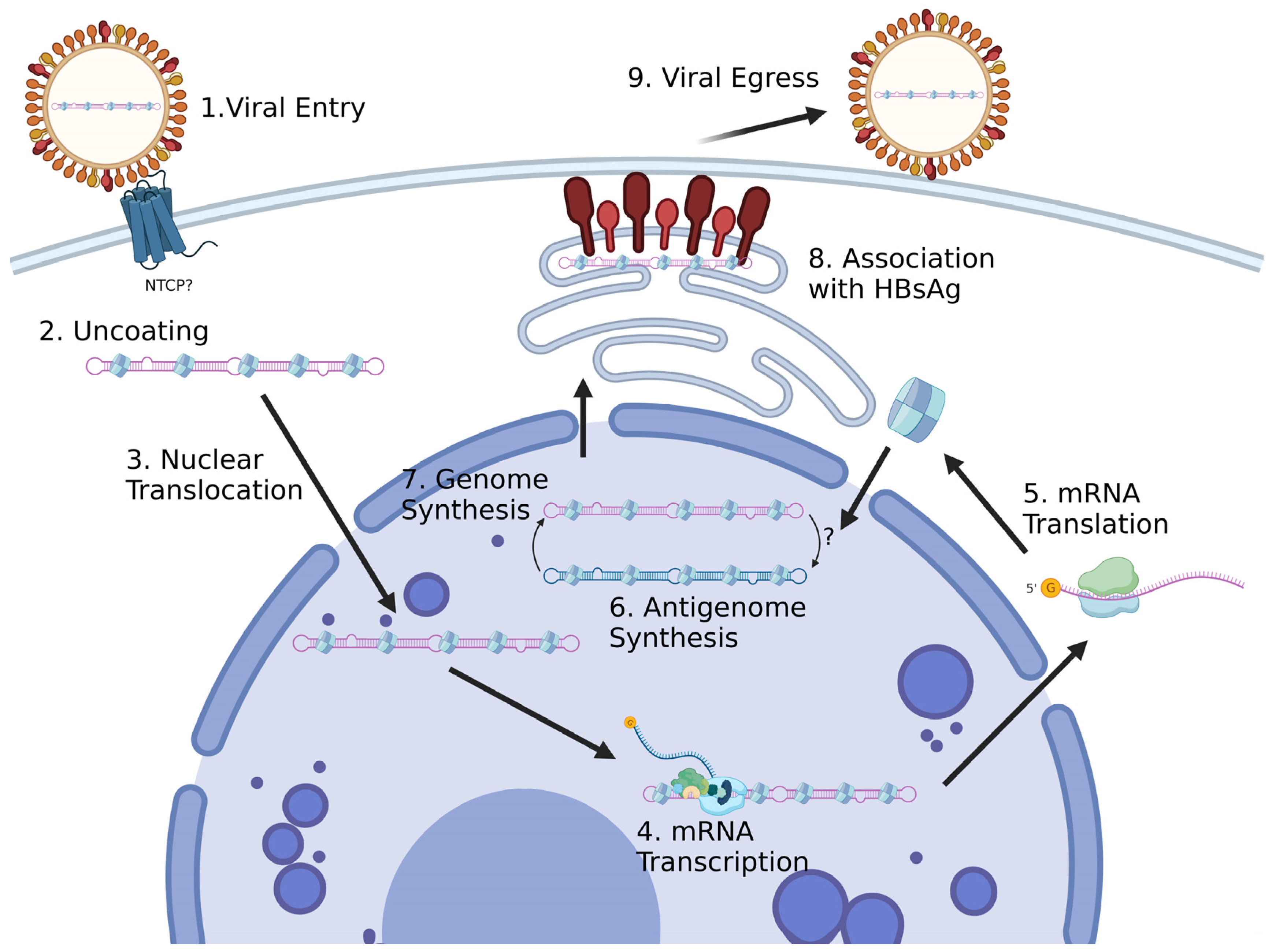
2.1. Viral Entry
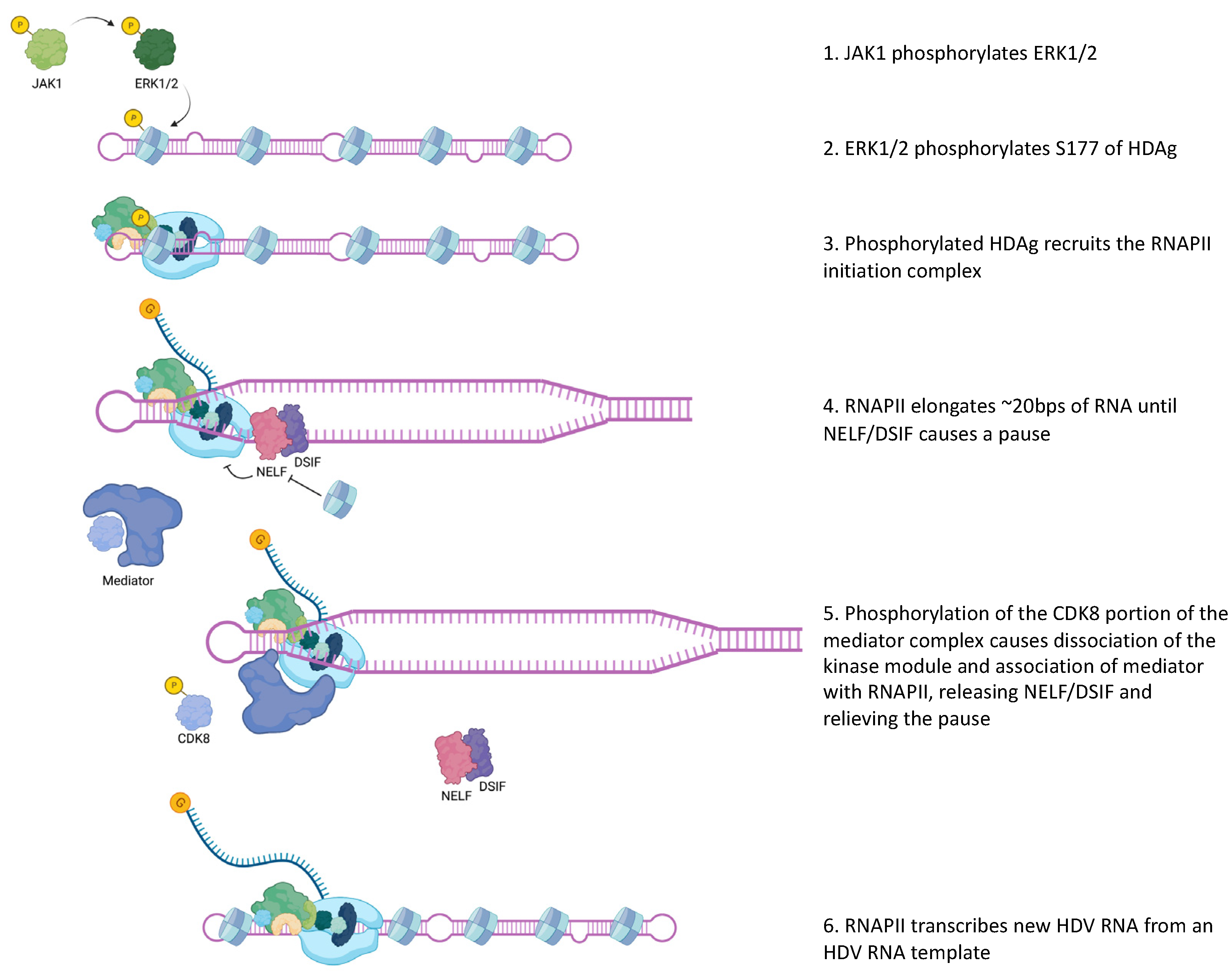
2.2. Viral Transcription
2.3. HDAg-S Functions
2.4. Viral Replication
2.5. HDAg-L Function and HDV Assembly
2.6. HDV Cellular Egress
2.7. Host Antiviral Activity
3. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taylor, J.M. Hepatitis D Virus Replication. Cold Spring Harb. Perspect. Med. 2015, 5, a021568. [Google Scholar] [CrossRef] [PubMed]
- Greco-Stewart, V.; Pelchat, M. Interaction of host cellular proteins with components of the hepatitis delta virus. Viruses 2010, 2, 189–212. [Google Scholar] [CrossRef] [PubMed]
- Goodrum, G.; Pelchat, M. Insight into the Contribution and Disruption of Host Processes during HDV Replication. Viruses 2018, 11, 21. [Google Scholar] [CrossRef]
- Lucifora, J.; Delphin, M. Current knowledge on Hepatitis Delta Virus replication. Antivir. Res. 2020, 179, 104812. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Netter, H.J.; Littlejohn, M.; Yuen, L.; Shi, M.; Eden, J.S.; Klaassen, M.; Holmes, E.C.; Hurt, A.C. A Divergent Hepatitis D-Like Agent in Birds. Viruses 2018, 10, 720. [Google Scholar] [CrossRef]
- Hetzel, U.; Szirovicza, L.; Smura, T.; Prahauser, B.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Identification of a Novel Deltavirus in Boa Constrictors. mBio 2019, 10, e00014-19. [Google Scholar] [CrossRef]
- Chang, W.S.; Pettersson, J.H.; Le Lay, C.; Shi, M.; Lo, N.; Wille, M.; Eden, J.S.; Holmes, E.C. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 2019, 5, vez021. [Google Scholar] [CrossRef]
- Paraskevopoulou, S.; Pirzer, F.; Goldmann, N.; Schmid, J.; Corman, V.M.; Gottula, L.T.; Schroeder, S.; Rasche, A.; Muth, D.; Drexler, J.F.; et al. Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl. Acad. Sci. USA 2020, 117, 17977–17983. [Google Scholar] [CrossRef]
- Park, J.H.; Iwamoto, M.; Yun, J.H.; Uchikubo-Kamo, T.; Son, D.; Jin, Z.; Yoshida, H.; Ohki, M.; Ishimoto, N.; Mizutani, K.; et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 2022, 606, 1027–1031. [Google Scholar] [CrossRef]
- Joshi, S.S.; Coffin, C.S. Hepatitis B virus lymphotropism: Emerging details and challenges. Biotechnol. Genet. Eng. Rev. 2018, 34, 139–151. [Google Scholar] [CrossRef]
- Perez-Vargas, J.; Amirache, F.; Boson, B.; Mialon, C.; Freitas, N.; Sureau, C.; Fusil, F.; Cosset, F.L. Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat. Commun. 2019, 10, 2098. [Google Scholar] [CrossRef]
- Moroianu, M.H.J.; Blobel, G.; Radu, A. Mammalian karyopherin a1B and a2B heterodimers: a1 or a2 subunit binds nuclear localization signal and B subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 1995, 92, 6532–6536. [Google Scholar] [CrossRef] [PubMed]
- Gudima, S.; Dingle, K.; Wu, T.T.; Moraleda, G.; Taylor, J. Characterization of the 5′ Ends for Polyadenylated RNAs Synthesized during the Replication of Hepatitis Delta Virus. J. Virol. 1999, 73, 6533–6539. [Google Scholar] [CrossRef]
- Gudima, S.; Wu, S.Y.; Chiang, C.M.; Moraleda, G.; Taylor, J. Origin of Hepatitis Delta Virus mRNA. J. Virol. 2000, 74, 7204–7210. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Cao, D.; Haussecker, D.; Huang, Y.; Kay, M.A. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA 2009, 15, 1971–1979. [Google Scholar] [CrossRef]
- Sikora, D.; Greco-Stewart, V.S.; Miron, P.; Pelchat, M. The hepatitis delta virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology 2009, 390, 71–78. [Google Scholar] [CrossRef]
- Abrahem, A.; Pelchat, M. Formation of an RNA polymerase II preinitiation complex on an RNA promoter derived from the hepatitis delta virus RNA genome. Nucleic Acids Res. 2008, 36, 5201–5211. [Google Scholar] [CrossRef]
- Mota, S.; Mendes, M.; Freitas, N.; Penque, D.; Coelho, A.V.; Cunha, C. Proteome analysis of a human liver carcinoma cell line stably expressing hepatitis delta virus ribonucleoproteins. J. Proteom. 2009, 72, 616–627. [Google Scholar] [CrossRef]
- Tavanez, J.P.; Caetano, R.; Branco, C.; Brito, I.M.; Miragaia-Pereira, A.; Vassilevskaia, T.; Quina, A.S.; Cunha, C. Hepatitis delta virus interacts with splicing factor SF3B155 and alters pre-mRNA splicing of cell cycle control genes. FEBS J. 2020, 287, 3719–3732. [Google Scholar] [CrossRef] [PubMed]
- Bichko, V.V.; Taylor, J.M. Redistribution of the Delta Antigens in Cells Replicating the Genome of Hepatitis Delta Virus. J. Virol. 1996, 70, 8064–8070. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.; Perez-Hernandez, D.; Vazquez, J.; Coelho, A.V.; Cunha, C. Proteomic changes in HEK-293 cells induced by hepatitis delta virus replication. J. Proteom. 2013, 89, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Heuschkel, M.J.; Bach, C.; Gerges, E.; Schuster, C.; Baumert, T.F.; Verrier, E.R. Presented at the International HBV Meeting 2022, Paris, France, 18–22 September 2022.
- Chen, Y.S.; Huang, W.H.; Hong, S.Y.; Tsay, Y.G.; Chen, P.J. ERK1/2-mediated phosphorylation of small hepatitis delta antigen at serine 177 enhances hepatitis delta virus antigenomic RNA replication. J. Virol. 2008, 82, 9345–9358. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.S.; Lo, S.J.; Chen, P.J.; Lee, Y.H. Casein Kinase II and Protein Kinase C Modulate Hepatitis Delta Virus RNA Replication by Not Empty Viral Particle Assembly. J. Virol. 1996, 70, 6190–6198. [Google Scholar] [CrossRef]
- Chen, C.W.; Tsay, Y.G.; Wu, H.L.; Lee, C.H.; Chen, D.S.; Chen, P.J. The double-stranded RNA-activated kinase, PKR, can phosphorylate hepatitis D virus small delta antigen at functional serine and threonine residues. J. Biol. Chem. 2002, 277, 33058–33067. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Stallcup, M.R.; Lai, M.M. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 2004, 78, 13325–13334. [Google Scholar] [CrossRef]
- Tseng, C.H.; Cheng, T.S.; Shu, C.Y.; Jeng, K.S.; Lai, M.M. Modification of small hepatitis delta virus antigen by SUMO protein. J. Virol. 2010, 84, 918–927. [Google Scholar] [CrossRef]
- Lee, C.Z.; Sheu, J.C. Histone H1e interacts with small hepatitis delta antigen and affects hepatitis delta virus replication. Virology 2008, 375, 197–204. [Google Scholar] [CrossRef]
- Huang, W.H.; Mai, R.T.; Lee, Y.H. Transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate hepatitis delta virus RNA replication. J. Virol. 2008, 82, 7313–7324. [Google Scholar] [CrossRef]
- Lee, C.H.; Chang, S.C.; Chen, C.J.; Chang, M.F. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 1998, 273, 7650–7656. [Google Scholar] [CrossRef]
- Huang, W.H.; Yung, B.Y.; Syu, W.J.; Lee, Y.H. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 2001, 276, 25166–25175. [Google Scholar] [CrossRef] [PubMed]
- Abeywickrama-Samarakoon, N.; Cortay, J.C.; Sureau, C.; Muller, S.; Alfaiate, D.; Guerrieri, F.; Chaikuad, A.; Schroder, M.; Merle, P.; Levrero, M.; et al. Hepatitis Delta Virus histone mimicry drives the recruitment of chromatin remodelers for viral RNA replication. Nat. Commun. 2020, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Greco-Stewart, V.S.; Miron, P.; Abrahem, A.; Pelchat, M. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virology 2007, 357, 68–78. [Google Scholar] [CrossRef]
- Lehmann, E.; Brueckner, F.; Cramer, P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 2007, 450, 445–449. [Google Scholar] [CrossRef]
- Stephenson-Tsoris, S.; Casey, J.L. Hepatitis Delta Virus Genome RNA Synthesis Initiates at Position 1646 with a Nontemplated Guanosine. J. Virol. 2022, 96, e02017–e02021. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Filipovska, J.; Yano, K.; Furuya, A.; Inukai, N.; Narita, T.; Wada, T.; Sugimoto, S.; Konarska, M.M.; Handa, H. Stimulation of RNA Polymerase II Elongation by Hepatitis Delta Antigen. Science 2001, 293, 124–127. [Google Scholar] [CrossRef]
- Urban, S. Presented at the Global Hepatitis Summit 2023, Paris, France, 25–28 April 2023.
- Chang, J.; Nie, X.; Chang, H.E.; Han, Z.; Taylor, J. Transcription of hepatitis delta virus RNA by RNA polymerase II. J. Virol. 2008, 82, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Greco-Stewart, V.S.; Schissel, E.; Pelchat, M. The hepatitis delta virus RNA genome interacts with the human RNA polymerases I and III. Virology 2009, 386, 12–15. [Google Scholar] [CrossRef]
- Li, Y.J.; Macnaughton, T.; Gao, L.; Lai, M.M. RNA-templated replication of hepatitis delta virus: Genomic and antigenomic RNAs associate with different nuclear bodies. J. Virol. 2006, 80, 6478–6486. [Google Scholar] [CrossRef]
- Modahl, L.E.; Macnaughton, T.B.; Zhu, N.; Johnson, D.L.; Lai, M.M. RNA-Dependant Replication and Transcription of Hepatitis Delta Virus RNA Involve Distinct Cellular RNA Polymerases. Mol. Cell. Biol. 2000, 20, 6030–6039. [Google Scholar] [CrossRef]
- Verrier, E.R.; Weiss, A.; Bach, C.; Heydmann, L.; Turon-Lagot, V.; Kopp, A.; El Saghire, H.; Crouchet, E.; Pessaux, P.; Garcia, T.; et al. Combined small molecule and loss-of-function screen uncovers estrogen receptor alpha and CAD as host factors for HDV infection and antiviral targets. Gut 2020, 69, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Greco-Stewart, V.S.; Thibault, C.S.; Pelchat, M. Binding of the polypyrimidine tract-binding protein-associated splicing factor (PSF) to the hepatitis delta virus RNA. Virology 2006, 356, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Beeharry, Y.; Goodrum, G.; Imperiale, C.J.; Pelchat, M. The Hepatitis Delta Virus accumulation requires paraspeckle components and affects NEAT1 level and PSP1 localization. Sci. Rep. 2018, 8, 6031. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef]
- Nohales, M.A.; Flores, R.; Daros, J.A. Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA 2012, 109, 13805–13810. [Google Scholar] [CrossRef]
- Yuan, Y.; Stumpf, F.M.; Schlor, L.A.; Schmidt, O.P.; Saumer, P.; Huber, L.B.; Frese, M.; Hollmuller, E.; Scheffner, M.; Stengel, F.; et al. Chemoproteomic discovery of a human RNA ligase. Nat. Commun. 2023, 14, 842. [Google Scholar] [CrossRef]
- Casey, J.L.; Gerin, J.L. Hepatitis D Virus RNA Editing: Specific Modification of Adenosine in the Antigenomic RNA. J. Virol. 1995, 69, 7593–7600. [Google Scholar] [CrossRef]
- Poison, A.G.; Bass, B.L.; Casey, J.L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 1996, 380, 454–456. [Google Scholar] [CrossRef]
- Jayan, G.C.; Casey, J.L. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 2002, 76, 3819–3827. [Google Scholar] [CrossRef]
- Wong, S.K.; Lazinski, D.W. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 2002, 99, 15118–15123. [Google Scholar] [CrossRef]
- Casey, J.L. Control of ADAR1 editing of hepatitis delta virus RNAs. Curr. Top Microbiol. Immunol. 2012, 353, 123–143. [Google Scholar] [CrossRef]
- Wang, Y.H.; Chang, S.C.; Huang, C.; Li, Y.P.; Lee, C.H.; Chang, M.F. Novel nuclear export signal-interacting protein, NESI, critical for the assembly of hepatitis delta virus. J. Virol. 2005, 79, 8113–8120. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Lee, C.P.; Liu, H.K.; Chang, M.F.; Lai, Y.H.; Lee, Y.C.; Huang, C. Cellular Nuclear Export Factors TAP and Aly Are Required for HDAg-L-mediated Assembly of Hepatitis Delta Virus. J. Biol. Chem. 2016, 291, 26226–26238. [Google Scholar] [CrossRef]
- Jeffrey, S.; Glenn, J.A.W.; Christopher, M. Havel and Judith, M. White. Identification of a Prenylation Site in Delta Virus Large Antigen. Science 1992, 256, 1331–1333. [Google Scholar]
- Chang, F.L.; Chen, P.J.; Tu, S.J.; Wang, C.J.; Chen, D.S. The large form of hepatitis d antigen is crucial for assembly of hepatitis d virus. Proc. Natl. Acad. Sci. USA 1991, 88, 8490–8494. [Google Scholar] [CrossRef] [PubMed]
- Bordier, B.B.; Ohkanda, J.; Liu, P.; Lee, S.-Y.; Salazar, F.H.; Marion, P.L.; Ohashi, K.; Meuse, L.; Kay, M.A.; Casey, J.L.; et al. In vivo antiviral efficacy of prenylation inhibitors against hepatitis delta virus. J. Clin. Investig. 2003, 112, 407–414. [Google Scholar] [CrossRef]
- Sureau, C.; Guerra, B.; Lanford, R.E. Role of the Large Hepatitis B Virus Envelope Protein in Infectivity of the Hepatitis Delta Virion. J. Virol. 1993, 67, 366–372. [Google Scholar] [CrossRef]
- Chao, M.; Hsieh, S.Y.; Taylor, J. Role of Two Forms of Hepatitis Delta Virus Antigen: Evidence for a Mechanism of Self-Limiting Genome Replication. J. Virol. 1990, 64, 5066–5069. [Google Scholar] [CrossRef]
- Modahl, L.E.; Lai, M.M. The Large Delta Antigen of Hepatitis Delta Virus Potently Inhibits Genomic by Not Antigenomic RNA Synthesis: A Mechanism Enabling Initiation of Viral Replication. J. Virol. 2000, 74, 7375–7380. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, M.; Li, S.; Bu, Y.; Xu, Z.; Zhu, G.; Wu, C.; Zhao, K.; Li, A.; Chen, Q.; et al. Hepatitis B virus hijacks TSG101 to facilitate egress via multiple vesicle bodies. PLoS Pathog. 2023, 19, e1011382. [Google Scholar] [CrossRef]
- Huang, C.; Chang, S.C.; Yu, I.C.; Tsay, Y.G.; Chang, M.F. Large hepatitis delta antigen is a novel clathrin adaptor-like protein. J. Virol. 2007, 81, 5985–5994. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Huang, C.R.; Chao, M.; Lo, S.J. The C-terminal sequence of the large hepatitis delta antigen is variable but retains the ability to bind clathrin. Virol. J. 2009, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Filzmayer, C.; Ni, Y.; Sultmann, H.; Mutz, P.; Hiet, M.S.; Vondran, F.W.R.; Bartenschlager, R.; Urban, S. Hepatitis D virus replication is sensed by MDA5 and induces IFN-beta/lambda responses in hepatocytes. J. Hepatol. 2018, 69, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Urban, S.; Neumann-Haefelin, C.; Lampertico, P. Hepatitis D virus in 2021: Virology, immunology and new treatment approaches for a difficult-to-treat disease. Gut 2021, 70, 1782–1794. [Google Scholar] [CrossRef]
- Gill, U.S. The immune landscape in hepatitis delta virus infection-Still an open field. J. Viral Hepat. 2023, 30 (Suppl. S1), 21–25. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stephenson-Tsoris, S.; Liang, T.J. Hepatitis Delta Virus–Host Protein Interactions: From Entry to Egress. Viruses 2023, 15, 1530. https://doi.org/10.3390/v15071530
Stephenson-Tsoris S, Liang TJ. Hepatitis Delta Virus–Host Protein Interactions: From Entry to Egress. Viruses. 2023; 15(7):1530. https://doi.org/10.3390/v15071530
Chicago/Turabian StyleStephenson-Tsoris, Susannah, and T. Jake Liang. 2023. "Hepatitis Delta Virus–Host Protein Interactions: From Entry to Egress" Viruses 15, no. 7: 1530. https://doi.org/10.3390/v15071530




