Emergence and Prevalence of an African Swine Fever Virus Variant in Wild Boar Populations in South Korea from 2019 to 2022
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of ASF Outbreak Distribution
2.2. DNA Extraction and ASFV Detection in Wild Boar Samples
2.3. Complete ASFV Genome Sequencing
2.4. ASFV Complete-Genome Annotation
2.5. Genetic ASFV Characterization and B646L (p72) Phylogenetic Analysis
3. Results
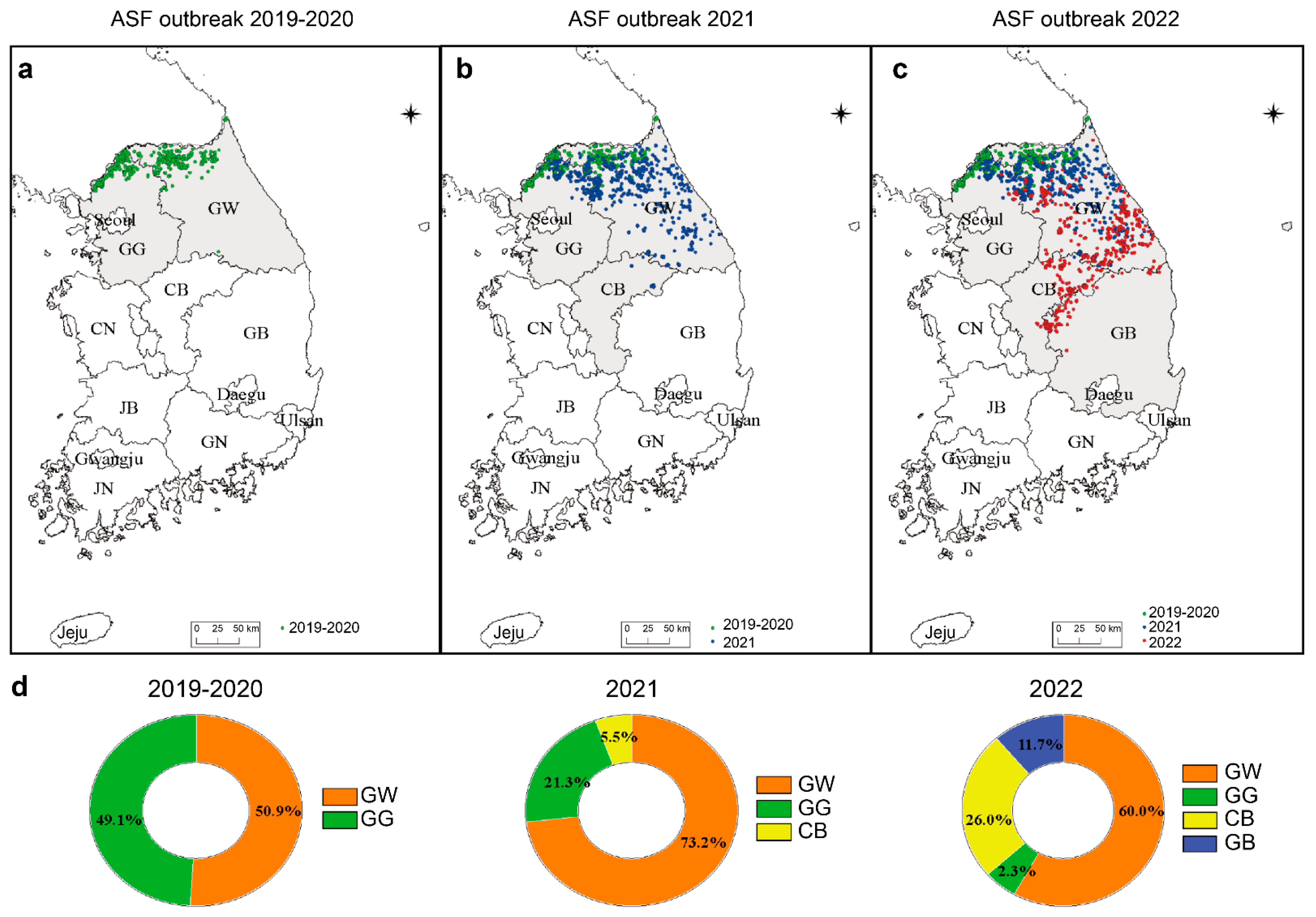
3.1. Geographical Distribution of ASF Outbreaks in South Korea during 2019–2022
3.2. Geographical Representation of the ASFV Genome Isolated from a Wild Boar with ASF in South Korea
3.3. Phylogenetic Analysis of Complete ASFV Strain Genomes
3.4. ASFV Genome Comparison of Korean Strains with Reported ASFV Strains
3.5. Amino Acid Sequence Alignment in the MGF 360-1La and 360-4L Regions
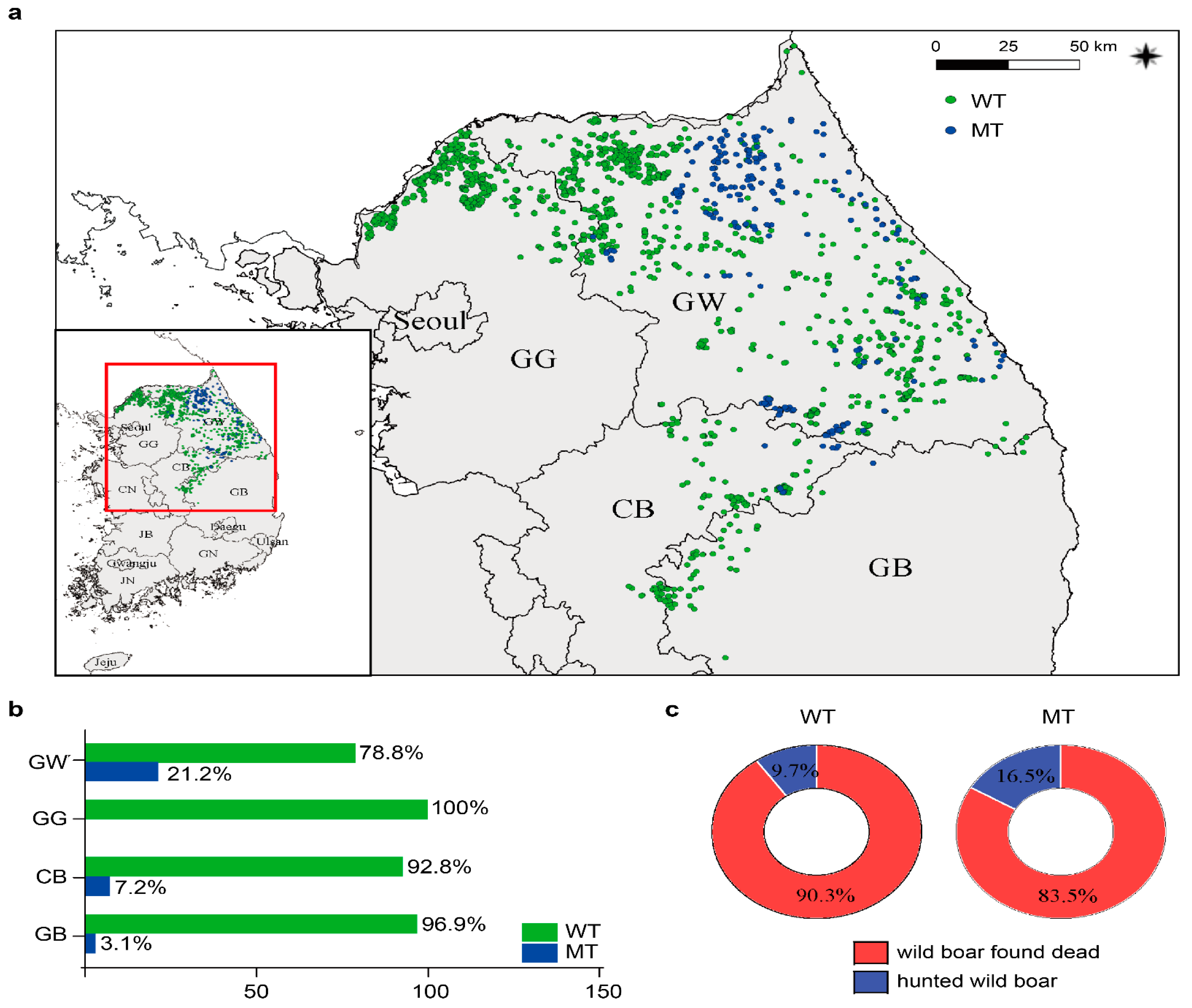
3.6. Geographic Distribution of Variant ASFV in Korea during 2019–2022
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salas, M.L. African Swine Fever Virus (Asfarviridae). In Encyclopedia of Virology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 30–38. [Google Scholar] [CrossRef]
- Costard, S.; Wieland, B.; De Glanville, W.; Jori, F.; Rowlands, R.; Vosloo, W.; Roger, F.; Pfeiffer, D.U.; Dixon, L.K. African swine fever: How can global spread be prevented? Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2683–2696. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Gomez-Villamandos, J.C.; Carrasco, L. An Update on the Epidemiology and Pathology of African Swine Fever. J. Comp. Pathol. 2015, 152, 9–21. [Google Scholar] [CrossRef]
- Penrith, M.-L. History of’swine fever in Southern Africa: Historical article. J. S. Afr. Vet. Assoc. 2013, 84, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Khomenko, S.; Beltrán-Alcrudo, D.; Rozstanlnyy, A.; Gogin, A.; Kolbasov, D.; Pinto, J.; Lubroth, J.; Martin, V. African Swine Fever in the Russian Federation: Risk Factors for Europe and Beyond. EMPRES Watch 2013, 28, 1–14. Available online: https://www.fao.org/publications/card/fr/c/456c5e1d-f450-524f-a664-/ (accessed on 25 July 2023).
- Gonzales, W.; Moreno, C.; Duran, U.; Henao, N.; Bencosme, M.; Lora, P.; Reyes, R.; Núñez, R.; De Gracia, A.; Perez, A.M. African swine fever in the Dominican Republic. Transbound. Emerg. Dis. 2021, 68, 3018–3019. [Google Scholar] [CrossRef]
- Kim, H.J.; Cho, K.H.; Lee, S.K.; Kim, D.Y.; Nah, J.J.; Kim, H.J.; Kim, H.J.; Hwang, J.Y.; Sohn, H.J.; Choi, J.G.; et al. Outbreak of African swine fever in South Korea, 2019. Transbound. Emerg. Dis. 2020, 67, 473–475. [Google Scholar] [CrossRef]
- Yoo, D.; Kim, H.; Lee, J.Y.; Yoo, H.S. African swine fever: Etiology, epidemiological status in Korea, and perspective on control. J. Vet. Sci. 2020, 21, e38. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.-S.; Andraud, M.; Kim, E.; Vergne, T. Three Years of African Swine Fever in South Korea (2019–2021): A Scoping Review of Epidemiological Un-derstanding. Transbound. Emerg. Dis. 2023, 2023, 4686980. [Google Scholar] [CrossRef]
- Karger, A.; Pérez-Núñez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.-F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef] [Green Version]
- Yáñez, R.J.; Rodriguez, J.M.; Nogal, M.L.; Yuste, L.; Enriquez, C.; Rodriguez, J.F.; Viñuela, E. Analysis of the Complete Nucleotide Sequence of African Swine Fever Virus. Virology 1995, 208, 249–278. [Google Scholar] [CrossRef] [Green Version]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Wen, X.; He, X.; Zhang, X.; Zhang, X.; Liu, L.; Guan, Y.; Zhang, Y.; Bu, Z. Genome sequences derived from pig and dried blood pig feed samples provide important insights into the transmission of African swine fever virus in China in 2018. Emerg. Microbes Infect. 2019, 8, 303–306. [Google Scholar] [CrossRef]
- Martins, C.; Boinas, F.S.; Iacolina, L.; Ruiz-Fons, F.; Gavier-Widén, D. African swine fever (ASF), the pig health challenge of the century. In Understanding and Combatting African Swine Fever: A European Perspective; Wageningen Academic Publishers: Wageningen, The Netherlands, 2021; pp. 149–154. [Google Scholar]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M.; ICTV Report Consortium. ICTV Virus Taxonomy Profile Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef]
- Bao, J.; Zhang, Y.; Shi, C.; Wang, Q.; Wang, S.; Wu, X.; Cao, S.; Xu, F.; Wang, Z. Genome-Wide Diversity Analysis of African Swine Fever Virus Based on a Curated Dataset. Animals 2022, 12, 2446. [Google Scholar] [CrossRef]
- Bastos, A.D.S.; Penrith, M.-L.; Crucière, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef]
- Achenbach, J.E.; Gallardo, C.; Nieto-Pelegrín, E.; Rivera-Arroyo, B.; Degefa-Negi, T.; Arias, M.; Jenberie, S.; Mulisa, D.D.; Gizaw, D.; Gelaye, E.; et al. Identification of a New Genotype of African Swine Fever Virus in Domestic Pigs from Ethiopia. Transbound. Emerg. Dis. 2017, 64, 1393–1404. [Google Scholar] [CrossRef]
- Li, X.; Xiao, K.; Zhang, Z.; Yang, J.; Wang, R.; Shen, X.; Pan, J.; Irwin, D.M.; Chen, R.-A.; Shen, Y. The recombination hot spots and genetic diversity of the genomes of African swine fever viruses. J. Infect. 2020, 80, 121–142. [Google Scholar] [CrossRef]
- Qu, H.; Ge, S.; Zhang, Y.; Wu, X.; Wang, Z. A systematic review of genotypes and serogroups of African swine fever virus. Virus Genes 2022, 58, 77–87. [Google Scholar] [CrossRef]
- Cackett, G.; Matelska, D.; Sýkora, M.; Portugal, R.; Malecki, M.; Bähler, J.; Dixon, L.; Werner, F. The African swine fever virus transcriptome. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Calvo, V.; Almazán, F.; Almendral, J.M.; Ramírez, J.C.; de la Vega, I.; Blasco, R.; Viñuela, E. Multigene families in African swine fever virus: Family 360. J. Virol. 1990, 64, 2073–2081. [Google Scholar] [CrossRef] [Green Version]
- Farlow, J.; Donduashvili, M.; Kokhreidze, M.; Kotorashvili, A.; Vepkhvadze, N.G.; Kotaria, N.; Gulbani, A. Intra-epidemic genome variation in highly pathogenic African swine fever virus (ASFV) from the country of Georgia. Virol. J. 2018, 15, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonso, C.L.; Piccone, M.E.; Zaffuto, K.M.; Neilan, J.; Kutish, G.F.; Lu, Z.; Balinsky, C.A.; Gibb, T.R.; Bean, T.J.; Zsak, L.; et al. African Swine Fever Virus Multigene Family 360 and 530 Genes Affect Host Interferon Response. J. Virol. 2004, 78, 1858–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazur-Panasiuk, N.; Woźniakowski, G.; Niemczuk, K. The first complete genomic sequences of African swine fever virus isolated in Poland. Sci. Rep. 2019, 9, 4556. [Google Scholar] [CrossRef] [Green Version]
- Senthilkumar, D.; Rajukumar, K.; Venkatesh, G.; Singh, F.; Tosh, C.; Kombiah, S.; Dubey, C.K.; Chakravarty, A.; Barman, N.N.; Singh, V.P. Complete genome analysis of African swine fever virus isolated from domestic pigs during the first ASF outbreaks in India. Transbound. Emerg. Dis. 2022, 69, e2020–e2027. [Google Scholar] [CrossRef] [PubMed]
- Forth, J.H.; Calvelage, S.; Fischer, M.; Hellert, J.; Sehl-Ewert, J.; Roszyk, H.; Deutschmann, P.; Reichold, A.; Lange, M.; Thulke, H.H.; et al. African swine fever virus–variants on the rise. Emerg. Microbes Infect. 2023, 12, 2146537. [Google Scholar] [CrossRef]
- Aguero, M.; Fernandez, J.; Romero, L.; Mascaraque, C.S.; Arias, M.; Sanchez-Vizcaino, J.M. Highly Sensitive PCR Assay for Routine Diagnosis of African Swine Fever Virus in Clinical Samples. J. Clin. Microbiol. 2003, 41, 4431–4434. [Google Scholar] [CrossRef] [Green Version]
- Tcherepanov, V.; Ehlers, A.; Upton, C. Genome Annotation Transfer Utility (GATU): Rapid annotation of viral genomes using a closely related reference genome. BMC Genom. 2006, 7, 150. [Google Scholar] [CrossRef]
- Forth, J.H.; Forth, L.F.; King, J.; Groza, O.; Hübner, A.; Olesen, A.S.; Höper, D.; Dixon, L.K.; Netherton, C.L.; Rasmussen, T.B.; et al. A Deep-Sequencing Workflow for the Fast and Efficient Generation of High-Quality African Swine Fever Virus Whole-Genome Sequences. Viruses 2019, 11, 846. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.; Park, J.-E.; Kim, S.-J.; Kim, Y.; Kim, W.; Kim, Y.-K.; Jheong, W. Complete genome analysis of the African swine fever virus isolated from a wild boar responsible for the first viral outbreak in Korea, 2019. Front. Veter. Sci. 2022, 9, 1080397. [Google Scholar] [CrossRef]
- Chapman, D.A.; Darby, A.C.; Da Silva, M.; Upton, C.; Radford, A.D.; Dixon, L.K. Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus. Emerg. Infect. Dis. 2011, 17, 599. [Google Scholar] [CrossRef]
- Jia, L.; Jiang, M.; Wu, K.; Hu, J.; Wang, Y.; Quan, W.; Hao, M.; Liu, H.; Wei, H.; Fan, W.; et al. Nanopore sequencing of African swine fever virus. Sci. China Life Sci. 2020, 63, 160–164. [Google Scholar] [CrossRef]
- Pikalo, J.; Schoder, M.E.; Sehl, J.; Breithaupt, A.; Tignon, M.; Cay, A.B.; Gager, A.M.; Fischer, M.; Beer, M.; Blome, S. The African swine fever virus isolate Belgium 2018/1 shows high virulence in European wild boar. Transbound. Emerg. Dis. 2020, 67, 1654–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Núñez, D.; Castillo-Rosa, E.; Vigara-Astillero, G.; García-Belmonte, R.; Gallardo, C.; Revilla, Y. Identification and Isolation of Two Different Subpopulations Within African Swine Fever Virus Arm/07 Stock. Vaccines 2020, 8, 625. [Google Scholar] [CrossRef] [PubMed]
- Njau, E.P.; Entfellner, J.-B.D.; Machuka, E.M.; Bochere, E.N.; Cleaveland, S.; Shirima, G.M.; Kusiluka, L.J.; Upton, C.; Bishop, R.P.; Pelle, R.; et al. The first genotype II African swine fever virus isolated in Africa provides insight into the current Eurasian pandemic. Sci. Rep. 2021, 11, 13081. [Google Scholar] [CrossRef]
- Bao, J.; Wang, Q.; Lin, P.; Liu, C.; Li, L.; Wu, X.; Chi, T.; Xu, T.; Ge, S.; Liu, Y.; et al. Genome comparison of African swine fever virus China/2018/Anhui XCGQ strain and related European p72 Genotype II strains. Transbound. Emerg. Dis. 2019, 66, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Pershin, A.; Shevchenko, I.; Igolkin, A.; Zhukov, I.; Mazloum, A.; Aronova, E.; Vlasova, N.; Shevtsov, A. A Long-Term Study of the Biological Properties of ASF Virus Isolates Originating from Various Regions of the Russian Federation in 2013–2018. Veter. Sci. 2019, 6, 99. [Google Scholar] [CrossRef] [Green Version]
- Zani, L.; Forth, J.H.; Forth, L.; Nurmoja, I.; Leidenberger, S.; Henke, J.; Carlson, J.; Breidenstein, C.; Viltrop, A.; Höper, D.; et al. Deletion at the 5′-end of Estonian ASFV strains associated with an attenuated phenotype. Sci. Rep. 2018, 8, 65. [Google Scholar] [CrossRef] [Green Version]
- Olasz, F.; Mészáros, I.; Marton, S.; Kaján, G.L.; Tamás, V.; Locsmándi, G.; Magyar, T.; Bálint, A.; Bányai, K.; Zádori, Z. A Simple Method for Sample Preparation to Facilitate Efficient Whole-Genome Sequencing of African Swine Fever Virus. Viruses 2019, 11, 1129. [Google Scholar] [CrossRef] [Green Version]
- Mazloum, A.; van Schalkwyk, A.; Shotin, A.; Igolkin, A.; Shevchenko, I.; Gruzdev, K.N.; Vlasova, N. Comparative Analysis of Full Genome Sequences of African Swine Fever Virus Isolates Taken from Wild Boars in Russia in 2019. Pathogens 2021, 10, 521. [Google Scholar] [CrossRef]
- Shi, K.; Liu, H.; Yin, Y.; Si, H.; Long, F.; Feng, S. Molecular Characterization of African Swine Fever Virus From 2019-2020 Outbreaks in Guangxi Province, Southern China. Front. Veter. Sci. 2022, 9, 912224. [Google Scholar] [CrossRef]
- de Villiers, E.P.; Gallardo, C.; Arias, M.; da Silva, M.; Upton, C.; Martin, R.; Bishop, R.P. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology 2010, 400, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Portugal, R.; Coelho, J.; Hoeper, D.; Little, N.S.; Smithson, C.; Upton, C.; Martins, C.; Leitão, A.; Keil, G.M. Related strains of African swine fever virus with different virulence: Genome comparison and analysis. J. Gen. Virol. 2015, 96, 408–419. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Chastagner, A.; Pereira De Oliveira, R.; Hutet, E.; Le Dimna, M.; Paboeuf, F.; Lucas, P.; Blanchard, Y.; Dixon, L.; Vial, L.; Le Potier, M.F. Coding-complete genome sequence of an African swine fever virus strain Liv13/33 isolate from ex-perimental transmission between pigs and Ornithodoros moubata ticks. Microbiol. Resour. Announc. 2020, 9, e00185-20. [Google Scholar] [CrossRef]
- Zsak, L.; Borca, M.V.; Risatti, G.R.; Zsak, A.; French, R.A.; Lu, Z.; Kutish, G.F.; Neilan, J.G.; Callahan, J.D.; Nelson, W.M.; et al. Preclinical Diagnosis of African Swine Fever in Contact-Exposed Swine by a Real-Time PCR Assay. J. Clin. Microbiol. 2005, 43, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, F.; Alcaraz, C.; Eiras, A.; Yáñez, R.J.; Rodriguez, J.M.; Alonso, C.; Escribano, J.M. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J. Virol. 1994, 68, 7244–7252. [Google Scholar] [CrossRef] [Green Version]
- Njau, E.P.; Machuka, E.M.; Cleaveland, S.; Shirima, G.M.; Kusiluka, L.J.; Okoth, E.A.; Pelle, R. African Swine Fever Virus (ASFV): Biology, Genomics and Genotypes Circulating in Sub-Saharan Africa. Viruses 2021, 13, 2285. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Vizcaíno, J.M.; Laddomada, A.; Arias, M.L. African swine fever virus. In Diseases of Swine, 11th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 443–452. [Google Scholar]
- Wang, L.; Luo, Y.; Zhao, Y.; Gao, G.F.; Bi, Y.; Qiu, H.J. Comparative genomic analysis reveals an ‘open’pan-genome of African swine fever virus. Transbound. Emerg. Dis. 2020, 67, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Blasco, R.; de la Vega, I.; Almazan, F.; Aguero, M.; Viñuela, E. Genetic variation of african swine fever virus: Variable regions near the ends of the viral DNA. Virology 1989, 173, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forth, J.H.; Tignon, M.; Cay, A.B.; Forth, L.F.; Höper, D.; Blome, S.; Beer, M. Comparative analysis of whole-genome sequence of African swine fever virus Belgium 2018/1. Emerg. Infect. Dis. 2019, 25, 1249. [Google Scholar] [CrossRef] [PubMed]
- Blasco, R.; Agüero, M.; Almendral, J.; Viñuela, E. Variable and constant regions in african swine fever virus DNA. Virology 1989, 168, 330–338. [Google Scholar] [CrossRef] [PubMed]
- He, W.-R.; Yuan, J.; Ma, Y.-H.; Zhao, C.-Y.; Yang, Z.-Y.; Zhang, Y.; Han, S.; Wan, B.; Zhang, G.-P. Modulation of Host Antiviral Innate Immunity by African Swine Fever Virus: A Review. Animals 2022, 12, 2935. [Google Scholar] [CrossRef]

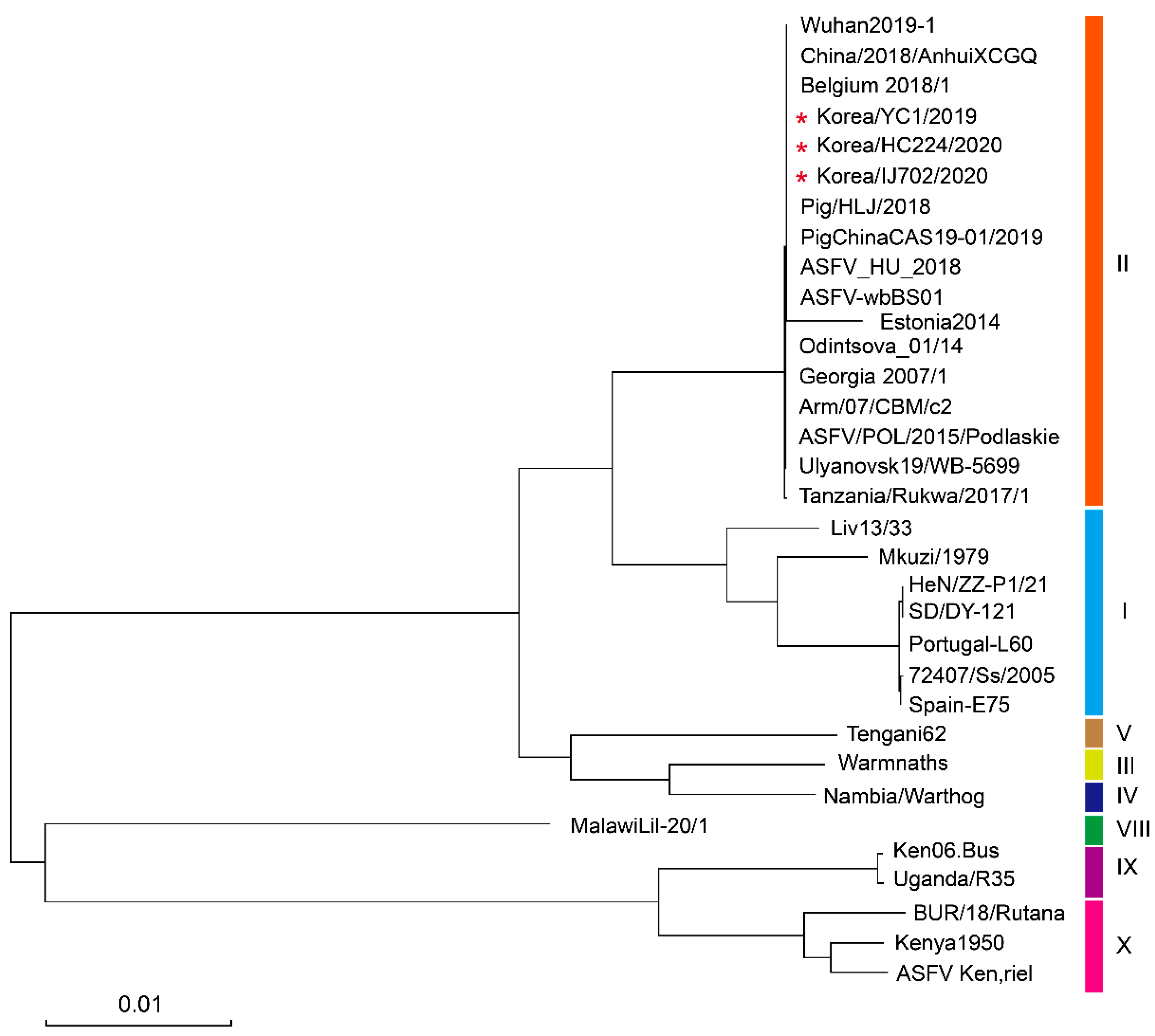

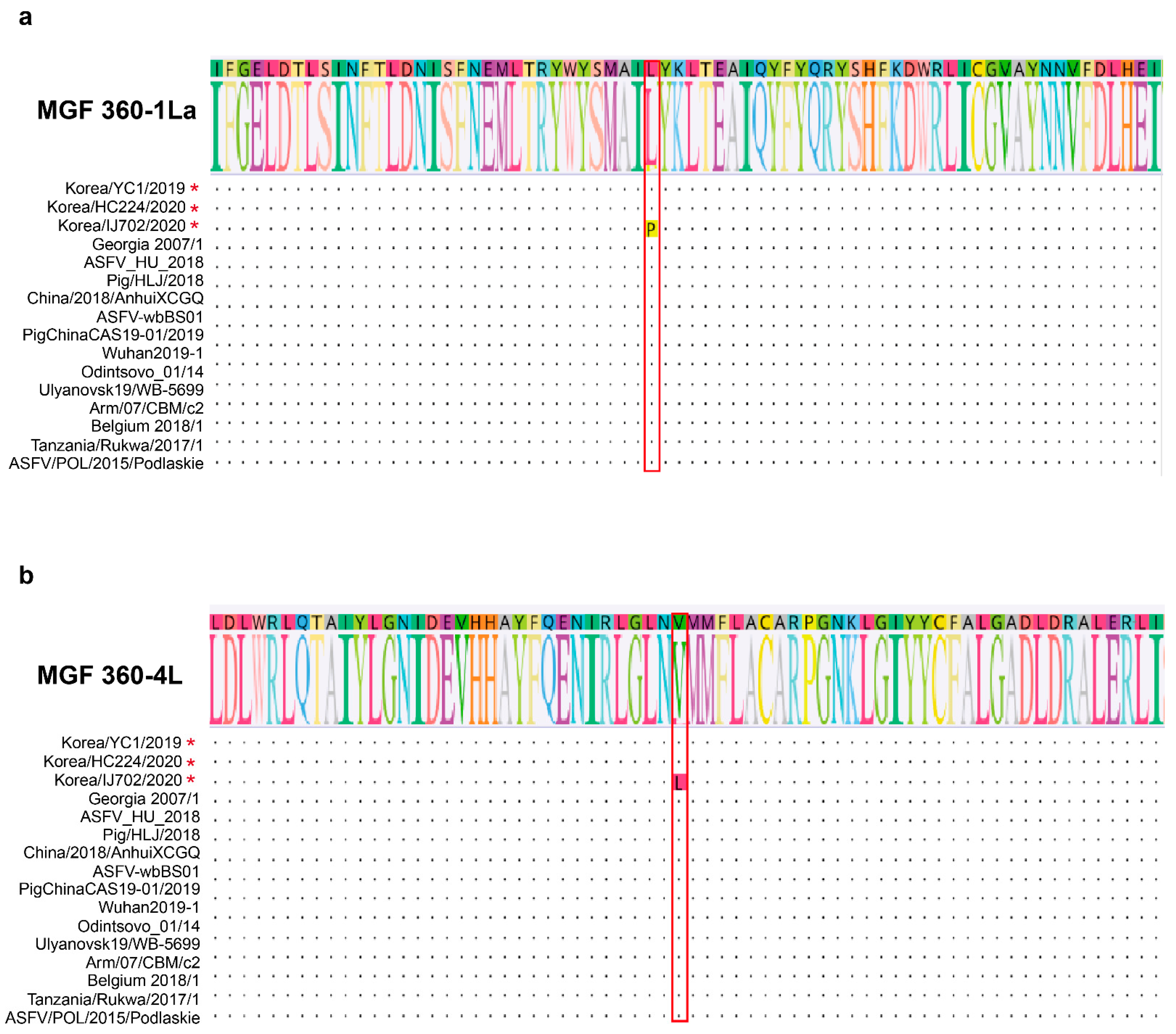
| Strain | Accession Number | Country | Year | Host | Length (bp) | Genotype | Ref. |
|---|---|---|---|---|---|---|---|
| Korea/YC1/2019 | ON075797 | Korea | 2019 | Wild boar | 188,950 | II | [31] |
| Korea/HC224/2020 | OP618183 | Korea | 2020 | Wild boar | 188,645 | II | Unpublished |
| Korea/IJ702/2020 | - | Korea | 2020 | Wild boar | 188,598 | II | Unpublished |
| Georgia 2007/1 | FR682468.2 | Georgia | 2007 | Pig | 190,584 | II | [32] |
| ASFV/pigChina/CAS19-01/2019 | MN172368 | China | 2019 | Pig | 189,405 | II | [33] |
| Belgium 2018/1 | LR536725 | Belgium | 2018 | Wild boar | 189,404 | II | [34] |
| ASFV-wbBS01 | MK645909 | China | 2018 | Wild boar | 189,394 | II | Unpublished |
| Arm/07/CBM/c2 | LR812933 | Armenia | 2007 | Pig | 190,145 | II | [35] |
| Wuhan2019-1 | MN393477 | China | 2019 | Pig | 190,576 | II | Unpublished |
| Tanzania/Rukwa/2017/1 | LR813622 | Tanzania | 2007 | Pig | 183,186 | II | [36] |
| China/2018/AnhuiXCGQ | MK128995 | China | 2018 | Pig | 189,393 | II | [37] |
| ASFV/POL/2015/Podlaskie | MH681419 | Poland | 2015 | Pig | 189,394 | II | [25] |
| Odintsovo_02/14 | KP843857 | Russia | 2014 | Wild boar | 189,333 | II | [38] |
| Estonia 2014 | LS478113 | Estonia | 2014 | Wild boar | 182,446 | II | [39] |
| ASFV_HU_2018 | MN715134 | China | 2018 | Pig | 190,601 | II | [40] |
| ASFV/Ulyanovsk 19/WB-5699 | MW306192 | Russia | 2019 | 189,263 | II | [41] | |
| Pig/HLJ/2018 | MK333180 | China | 2018 | Pig | 189,404 | II | [42] |
| Spain-E75 | NC_044958.1 | Spain | 181,187 | I | [43] | ||
| Portugal-L60 | NC_044941 | Portugal | 1960 | 182,362 | I | [44] | |
| HeN/ZZ-P1/21 | MZ945536 | China | 2021 | Pig | 171,235 | I | [45] |
| SD/DY-1-21 | MZ945537 | China | 2021 | Pig | 172,025 | I | [45] |
| 72407/Ss/2005 | MN270978 | Italy | 2005 | Sus scrofa | 181,699 | I | Unpublished |
| Mkuzi/1979 | AY261362 | South Africa | 1979 | 192,714 | I | Unpublished | |
| Liv13/33 | MN913970 | Zambia | 2017 | Ornithodoros moubata | 188,277 | I | [46] |
| Warmbaths | AY261365 | South Africa | 1987 | Tick | 190,773 | III | Unpublished |
| Namibia/Warthog | AY261366 | Namibia | 1980 | Warthog | 186,5258 | IV | [47] |
| Ken06.Bus | KM111295 | Kenya | 2006 | Pig | 184,368 | IX | Unpublished |
| Uganda/R35 | MH025920 | Uganda | 2015 | Pig | 188,629 | IX | Unpublished |
| Tengani62 | AY261364 | Malawi | 1962 | Pig | 185,689 | V | Unpublished |
| MalawiLil-20/1 | AY261361 | Malawi | 1983 | Tick | 187,162 | VIII | [48] |
| ASFV Ken.rie1 | LR899131 | Kenya | 2019 | Tick | 190,592 | X | Unpublished |
| Kenya1950 | AY261360 | Kenya | 1950 | Pig | 193,886 | X | [49] |
| BUR/18/Rutana | MW856067 | Burundi | 2018 | Pig | 176,564 | X | [50] |
| Name | Accession Number | Nonsynonymous | Synonymous | |
|---|---|---|---|---|
| MGF 360-1La | MGF360-4L | MGF 360-10L | ||
| Korea/YC1/2019 | ON075797 | A | C | G |
| Korea/HC224/2020 | OP618183 | A | C | G |
| Korea/IJ702/2020 | - | G | G | C |
| Georgia2007 | FR682468 | A | C | C |
| HU2018 | MN715134 | A | C | C |
| Pig/HLJ | MK333180 | A | C | C |
| AnhuiXCGQ/2018 | MK128995 | A | C | C |
| wbBS01 | MK645909 | A | C | C |
| Pig/china2019 | MN172368 | A | C | C |
| Wuhan2019 | MN393477 | A | C | C |
| Odintsovo | KP843857 | A | C | C |
| ulyanovsk19 | MW306192.1 | A | C | C |
| Arm/07/CBM | LR812933 | A | C | C |
| Belgium2018 | LR536725 | A | C | C |
| Tanzania2017 | LR813622 | A | C | C |
| Podlaskie2015 | MH681419 | A | C | C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, G.; Kim, S.-J.; Kim, W.-J.; Kim, J.-H.; Kim, J.-C.; Lee, S.-G.; Kim, E.-S.; Lee, S.-H.; Jheong, W.-H. Emergence and Prevalence of an African Swine Fever Virus Variant in Wild Boar Populations in South Korea from 2019 to 2022. Viruses 2023, 15, 1667. https://doi.org/10.3390/v15081667
Kim G, Kim S-J, Kim W-J, Kim J-H, Kim J-C, Lee S-G, Kim E-S, Lee S-H, Jheong W-H. Emergence and Prevalence of an African Swine Fever Virus Variant in Wild Boar Populations in South Korea from 2019 to 2022. Viruses. 2023; 15(8):1667. https://doi.org/10.3390/v15081667
Chicago/Turabian StyleKim, Garam, So-Jeong Kim, Won-Jun Kim, Jung-Hyeuk Kim, Ji-Chul Kim, Sang-Geon Lee, Eun-Sol Kim, Sang-Hyun Lee, and Weon-Hwa Jheong. 2023. "Emergence and Prevalence of an African Swine Fever Virus Variant in Wild Boar Populations in South Korea from 2019 to 2022" Viruses 15, no. 8: 1667. https://doi.org/10.3390/v15081667
APA StyleKim, G., Kim, S.-J., Kim, W.-J., Kim, J.-H., Kim, J.-C., Lee, S.-G., Kim, E.-S., Lee, S.-H., & Jheong, W.-H. (2023). Emergence and Prevalence of an African Swine Fever Virus Variant in Wild Boar Populations in South Korea from 2019 to 2022. Viruses, 15(8), 1667. https://doi.org/10.3390/v15081667






