Geobacillus Bacteriophages from Compost Heaps: Representatives of Three New Genera within Thermophilic Siphoviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Phages and Bacterial Strains
2.2. Phage Isolation, Propagation, and Purification Techniques
2.3. Transmission Electron Microscopy
2.4. DNA Isolation
2.5. Genome Sequencing and Analysis
2.6. Analysis of Structural Proteins
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Identification and Characterization of the Bacterial Isolates
3.2. Host Range, Morphology, and Physiological Characteristics of the Phages
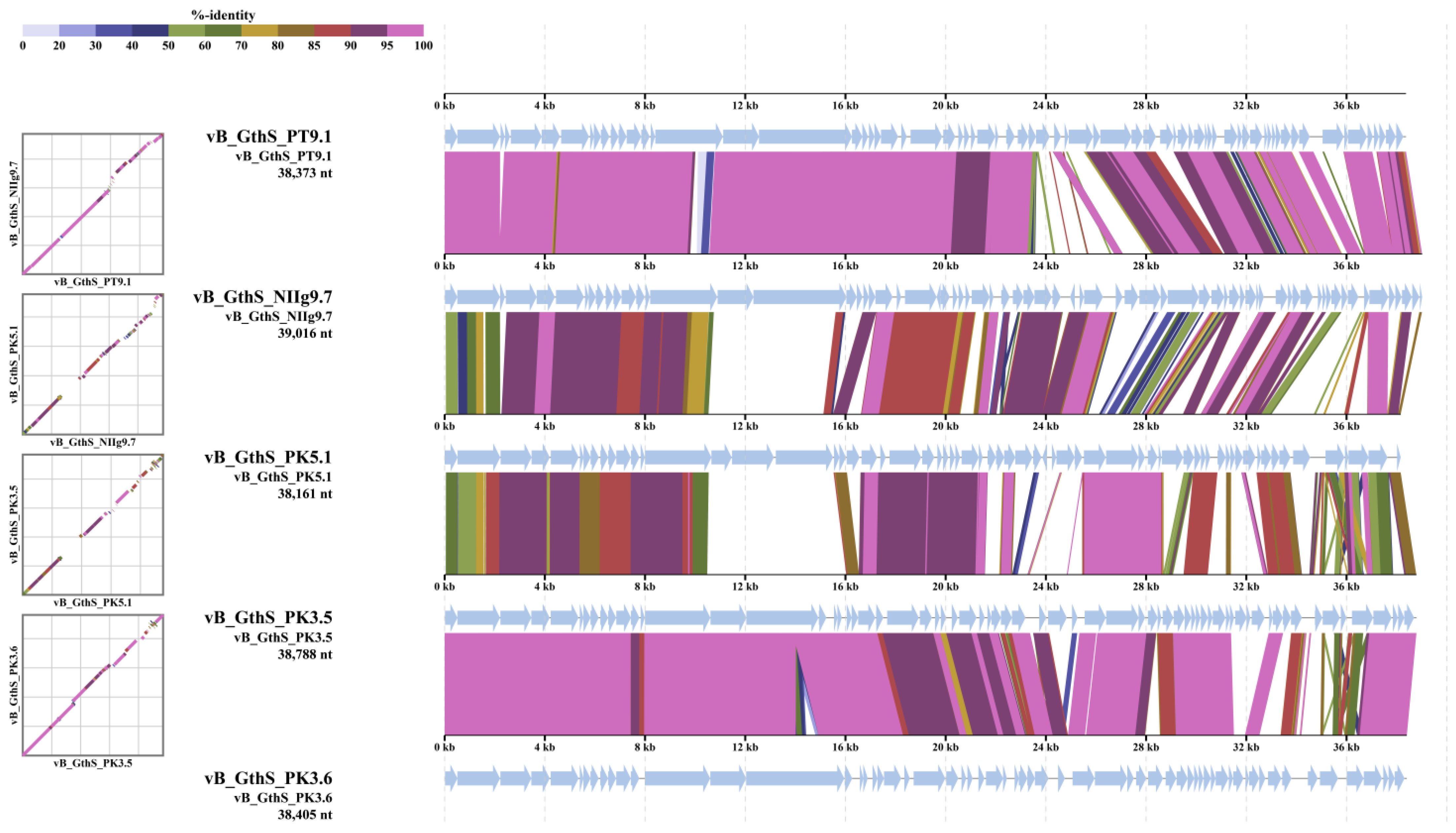
3.3. Overview of Viral Genomes
3.4. Structural Proteins and Proteomic Analysis
3.5. Packaging
3.6. DNA Replication, Recombination, and Repair
3.7. Transcription, Translation, Nucleotide Metabolism, and DNA Modification
3.8. Lysis Cassette
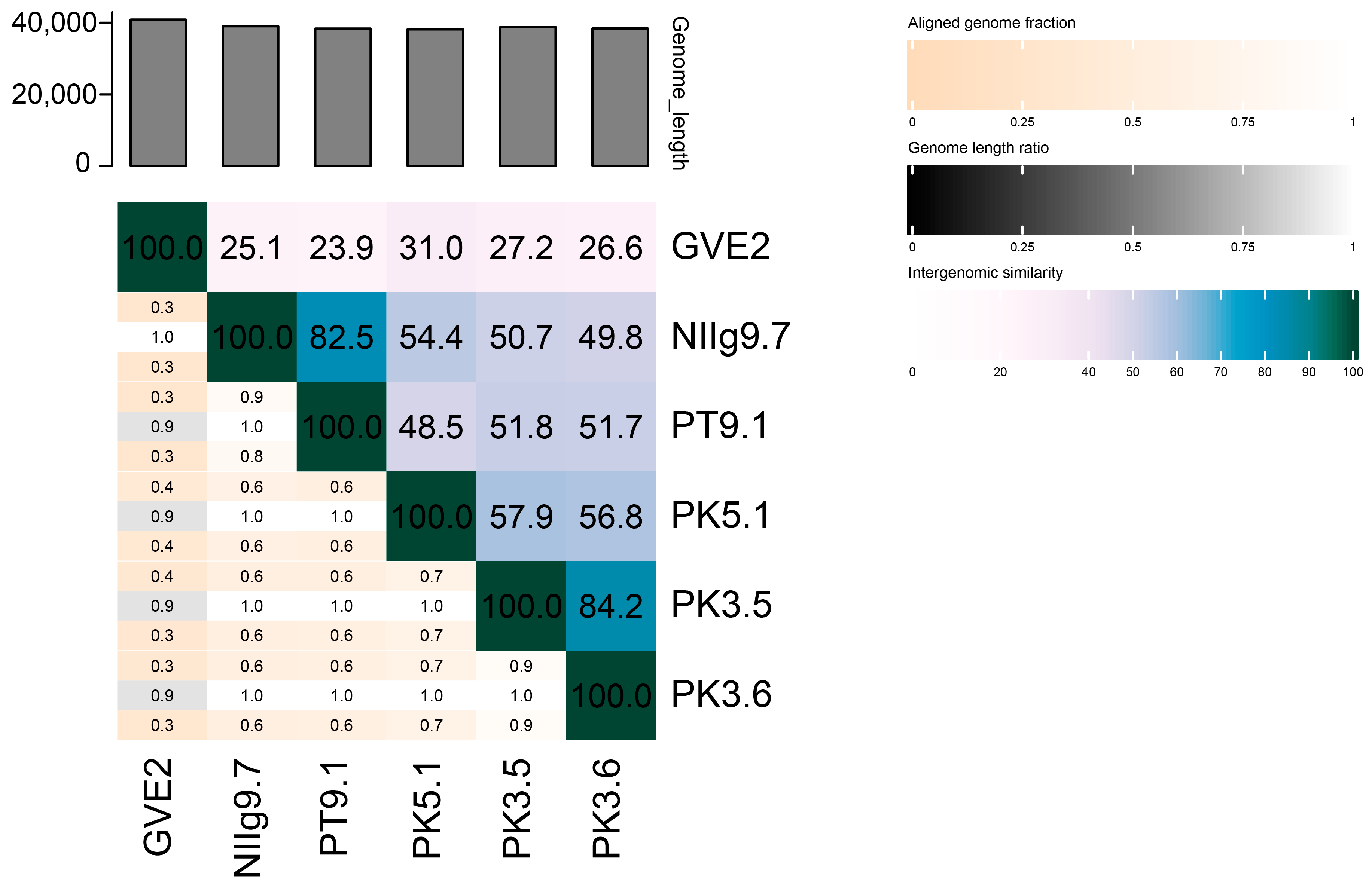
3.9. Phylogenetic Analysis of the Phages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McMullan, G.; Christie, J.M.; Rahman, T.J.; Banat, I.M.; Ternan, N.G.; Marchant, R. Habitat, applications and genomics of the aerobic, thermophilic genus Geobacillus. Biochem. Soc. Trans. 2004, 32, 214–217. [Google Scholar] [CrossRef]
- Najar, I.N.; Thakur, N. A systematic review of the genera Geobacillus and Parageobacillus: Their evolution, current taxonomic status and major applications. Microbiology 2020, 166, 800–816. [Google Scholar] [CrossRef]
- Zeigler, D.R. The Geobacillus paradox: Why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology 2014, 160, 1–11. [Google Scholar] [CrossRef]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. The genus Geobacillus and their biotechnological potential. Adv. Appl. Microbiol. 2015, 92, 1–48. [Google Scholar]
- Wada, K.; Suzuki, H. Chapter 15 Biotechnological platforms of the moderate thermophiles, Geobacillus species: Notable properties and genetic tools. In Physiological and Biotechnological Aspects of Extremophiles; Salwan, R., Sharma, V., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 195–218. [Google Scholar]
- Zebrowska, J.; Witkowska, M.; Struck, A.; Laszuk, P.E.; Raczuk, E.; Ponikowska, M.; Skowron, P.M.; Zylicz-Stachula, A. Antimicrobial potential of the genera Geobacillus and Parageobacillus, as well as endolysins biosynthesized by their bacteriophages. Antibiotics 2022, 11, 242. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, X. Deep-sea thermophilic Geobacillus bacteriophage GVE2 transcriptional profile and proteomic characterization of virions. Appl. Microbiol. Biotechnol. 2008, 80, 697–707. [Google Scholar] [CrossRef]
- van Zyl, L.J.; Sunda, F.; Taylor, M.P.; Cowan, D.; Trindade, M.I. Identification and characterization of a novel Geobacillus thermoglucosidasius bacteriophage, GVE3. Arch. Virol. 2015, 160, 2269–2282. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X. Genome analysis of deep-sea thermophilic bacteriophage D6E. Appl. Environ. Microbiol. 2010, 76, 7861–7866. [Google Scholar] [CrossRef]
- Marks, T.J.; Hamilton, P.T. Characterization of a thermophilic bacteriophage of Geobacillus kaustophilus. Arch. Virol. 2014, 159, 2771–2775. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, F.; Wu, S.; Xu, Y.; Zhang, X. Genomic and proteomic characterization of a thermophilic Geobacillus bacteriophage GBSV1. Res. Microbiol. 2009, 16, 166–171. [Google Scholar] [CrossRef]
- Liu, B.; Wu, S.; Xie, L. Complete genome sequence and proteomic analysis of a thermophilic bacteriophage BV1. Acta Oceanol. Sin. 2010, 29, 84–89. [Google Scholar] [CrossRef]
- Doi, K.; Mori, K.; Martono, H.; Nagayoshi, Y.; Fujino, Y.; Tashiro, K.; Kuhara, S.; Ohshima, T. Draft genome sequence of Geobacillus kaustophilus GBlys, a lysogenic strain with bacteriophage φOH2. Genome Announc. 2013, 1, e00634-13. [Google Scholar] [CrossRef]
- Skowron, P.M.; Kropinski, A.M.; Zebrowska, J.; Janus, L.; Szemiako, K.; Czajkowska, E.; Maciejewska, N.; Skowron, M.; Łoś, J.; Łoś, M.; et al. Sequence, genome organization, annotation and proteomics of the thermophilic, 47.7-kb Geobacillus stea-rothermophilus bacteriophage TP-84 and its classification in the new Tp84virus genus. PLoS ONE 2018, 13, e0195449. [Google Scholar]
- Choi, D.; Kong, M. LysGR1, a novel thermostable endolysin from Geobacillus stearothermophilus bacteriophage GR1. Front. Microbiol. 2023, 14, 1178748. [Google Scholar] [CrossRef]
- Liu, B.; Wu, S.; Song, Q.; Zhang, X.; Xie, L. Two novel bacteriophages of thermophilic bacteria isolated from deep-sea hydrothermal fields. Curr. Microbiol. 2006, 53, 163–166. [Google Scholar] [CrossRef]
- Łubkowska, B.; Jeżewska-Frąckowiak, J.; Sobolewski, I.; Skowron, P.M. Bacteriophages of thermophilic ‘Bacillus group’ bacteria—A review. Microorganisms 2021, 9, 1522. [Google Scholar] [CrossRef]
- Godon, J.J.; Zumstein, E.; Dabert, P.; Habouzit, F.; Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997, 63, 2802–2813. [Google Scholar] [CrossRef]
- Versalovic, J.; Schneider, M.; de Bruijn, F.J.; Lupski, J.R. Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR). Methods Cell. Mol. Biol. 1994, 5, 25–40. [Google Scholar]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Šimoliūnas, E.; Kaliniene, L.; Stasilo, M.; Truncaitė, L.; Zajančkauskaitė, A.; Staniulis, J.; Nainys, J.; Kaupinis, A.; Valius, M.; Meškys, R. Isolation and characterization of vB_ArS-ArV2—First Arthrobacter sp. infecting bacteriophage with completely sequenced genome. PLoS ONE 2014, 9, e111230. [Google Scholar] [CrossRef]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Šimoliūnas, E.; Kaliniene, L.; Truncaitė, L.; Zajančkauskaitė, A.; Staniulis, J.; Kaupinis, J.; Ger, M.; Valius, M.; Meškys, R. Klebsiella phage vB_KleM-RaK2—A giant singleton virus of the family Myoviridae. PLoS ONE 2013, 8, e60717. [Google Scholar] [CrossRef]
- Carlson, K.; Miller, E. Experiments in T4 genetics. In Bacteriophage T4; Karam, J.D., Ed.; ASM Press: Washington, DC, USA, 1994; pp. 419–483. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 11 July 2023).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Nikolenko, S.I.; Korobeynikov, A.I.; Alekseyev, M.A. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genom. 2013, 14, S7. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Alva, V.; Nam, S.Z.; Söding, J.; Lupas, A.N. The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016, 44, W410–W415. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented mpi bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. Viptree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Meintanis, C.; Chalkou, K.I.; Kormas, K.A.; Lymperopoulou, D.S.; Katsifas, E.A.; Hatzinikolaou, D.G.; Karagouni, A.D. Application of rpoB sequence similarity analysis, REP-PCR and BOX-PCR for the differentiation of species within the genus Geobacillus. Lett. Appl. Microbiol. 2008, 46, 395–401. [Google Scholar] [CrossRef]
- Bradley, D.E. Ultrastructure of bacteriophages and bacteriocins. Bacteriol. Rev. 1967, 31, 230–314. [Google Scholar] [CrossRef]
- Ackermann, H.W.; Eisenstark, A. The present state of phage taxonomy. Intervirology 1974, 3, 201–219. [Google Scholar] [CrossRef]
- Aliyu, H.; Lebre, P.; Blom, J.; Cowan, D.; De Maayer, P. Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 2016, 39, 527–533. [Google Scholar] [CrossRef]
- Rao, V.B.; Feiss, M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008, 42, 647–681. [Google Scholar] [CrossRef]
- Rao, V.B.; Feiss, M. Mechanisms of DNA packaging by large double stranded DNA viruses. Annu. Rev. Virol. 2015, 2, 351–378. [Google Scholar] [CrossRef]
- Edgell, D.R.; Gibb, E.A.; Belfort, M. Mobile DNA elements in T4 and related phages. Virol. J. 2010, 7, 290. [Google Scholar] [CrossRef]
- Young, I.; Wang, I.; Roof, W.D. Phages will out: Strategies of host cell lysis. Trends Microbiol. 2000, 8, 120–128. [Google Scholar] [CrossRef]
- Cowles, K.N.; Goodrich-Blair, H. Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell. Microbiol. 2005, 7, 209–219. [Google Scholar] [CrossRef]
- Fernandes, S.; São-José, C. Probing the function of the two holin-like proteins of bacteriophage SPP1. Virology 2017, 500, 184–189. [Google Scholar] [CrossRef]
- Halgasova, N.; Ugorcakova, J.; Gerova, M.; Timko, J.; Bukovska, G. Isolation and characterization of bacteriophage PhiBP from Paenibacillus polymyxa CCM 7400. FEMS Microbiol. Lett. 2010, 305, 128–135. [Google Scholar] [CrossRef]
- Nakonieczna, A.; Rutyna, P.; Fedorowicz, M.; Kwiatek, M.; Mizak, L.; Łobocka, M. Three novel bacteriophages, J5a, F16Ba, and z1a, specific for Bacillus anthracis, define a new clade of historical Wbeta phage relatives. Viruses 2022, 14, 213. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Cowan, D.A. Using signature genes as tools to assess environmental viral ecology and diversity. Appl. Environ. Microbiol. 2014, 80, 4470–4480. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A roadmap for genome-based phage taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Wells-Bennik, M.H.J.; Janssen, P.W.M.; Klaus, V.; Yang, C.; Zwietering, M.H.; den Besten, H.M.W. Heat resistance of spores of 18 strains of Geobacillus stearothermophilus and impact of culturing conditions. Int. J. Food Microbiol. 2019, 291, 161–172. [Google Scholar] [CrossRef]
- Andre, S.; Zuber, F.; Remize, F. Thermophilic spore-forming bacteria isolated from spoiled canned food and their heat resistance. Results of a French ten-year survey. Int. J. Food Microbiol. 2013, 165, 134–143. [Google Scholar] [CrossRef]
- Sadiq, F.A.; Flint, S.; Yuan, L.; Li, Y.; Liu, T.J.; He, G.Q. Propensity for biofilm formation by aerobic mesophilic and thermophilic spore forming bacteria isolated from chinese milk powders. Int. J. Food Microbiol. 2017, 262, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Karaca, B.; Buzrul, S.; Cihan, A.C. Anoxybacillus and Geobacillus biofilms in the dairy industry: Effects of surface material, incubation temperature and milk type. Biofouling 2019, 35, 551–560. [Google Scholar] [CrossRef]
- Ye, T.; Zhang, X. Characterization of a lysin from deep-sea thermophilic bacteriophage GVE2. Appl. Microbiol. Biotechnol. 2008, 78, 635–641. [Google Scholar] [CrossRef]
- Jin, M.; Ye, T.; Zhang, X. Roles of bacteriophage GVE2 endolysin in host lysis at high temperatures. Microbiology 2013, 159, 1597–1605. [Google Scholar] [CrossRef]
- Swift, S.M.; Reid, K.P.; Donovan, D.M.; Ramsay, T.G. Thermophile lytic enzyme fusion proteins that target Clostridium perfringens. Antibiotics 2019, 8, 214. [Google Scholar] [CrossRef]
- Miller, E.; Warek, U.; Xu, D. Endolysin from Bacteriophage against Geobacillus and Methods of Using. International Patent Application WIPO/PCT WO 2016/123425 Al, 4 August 2016. [Google Scholar]
- Żebrowska, J.; Żołnierkiewicz, O.; Ponikowska, M.; Puchalski, M.; Krawczun, N.; Makowska, J.; Skowron, P. Cloning and characterization of a thermostable endolysin of bacteriophage TP-84 as a potential disinfectant and biofilm-removing biological agent. Int. J. Mol. Sci. 2022, 23, 7612. [Google Scholar] [CrossRef] [PubMed]
- Łubkowska, B.; Czajkowska, E.; Stodolna, A.; Sroczyński, M.; Zylicz-Stachula, A.; Ireneusz, S.; Skowron, M.P. A novel thermostable TP-84 capsule depolymerase: A method for rapid polyethyleneimine processing of a bacteriophage-expressed proteins. Microb. Cell Factories 2023, 22, 80. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, A.; Ceyssens, P.J.; Krylov, V.N.; Noben, J.P.; Volckaert, G.; Lavigne, R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 2012, 434, 251–256. [Google Scholar] [CrossRef]
- Harper, D.; Parracho, H.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and function of phage encoded depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar]

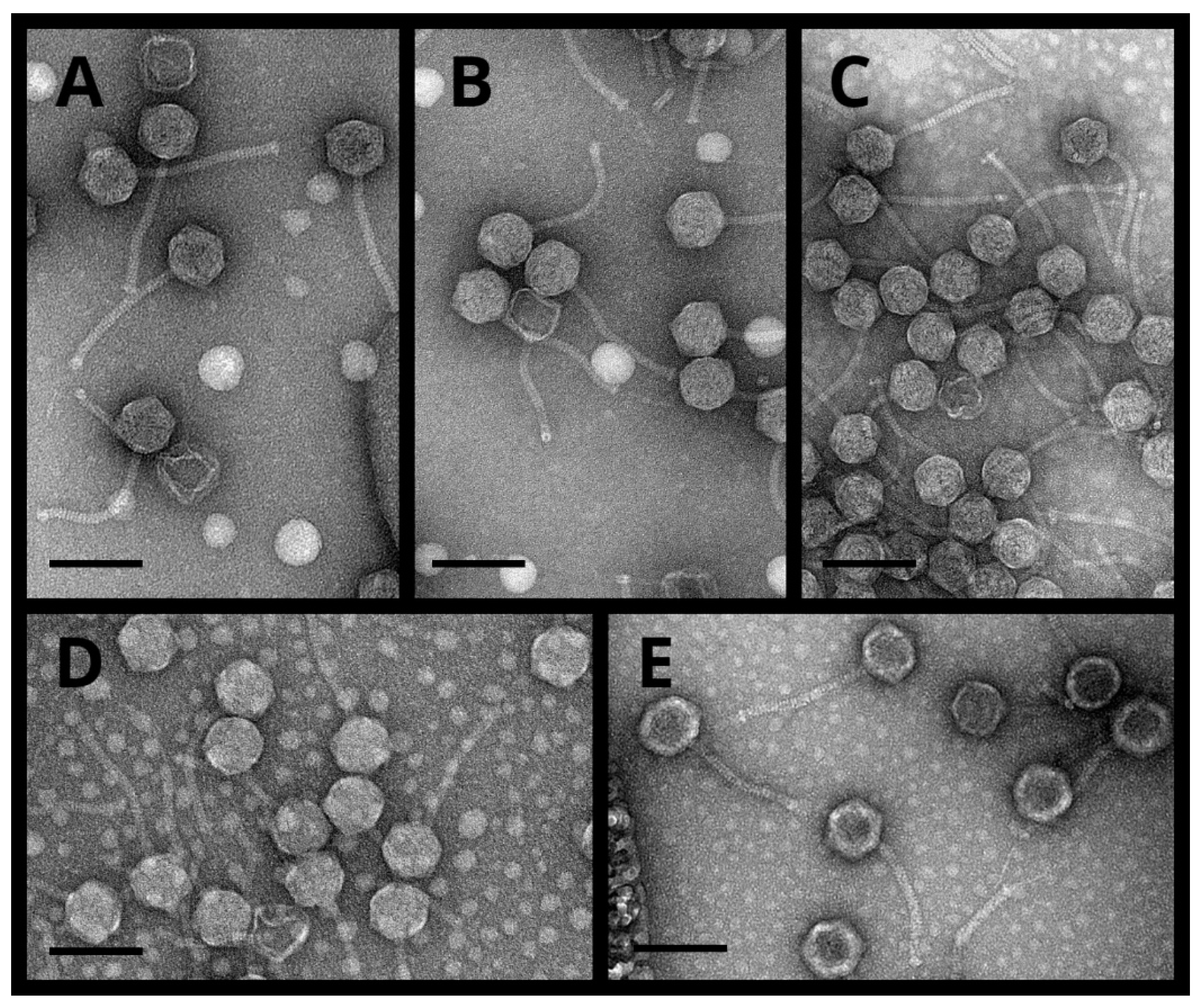
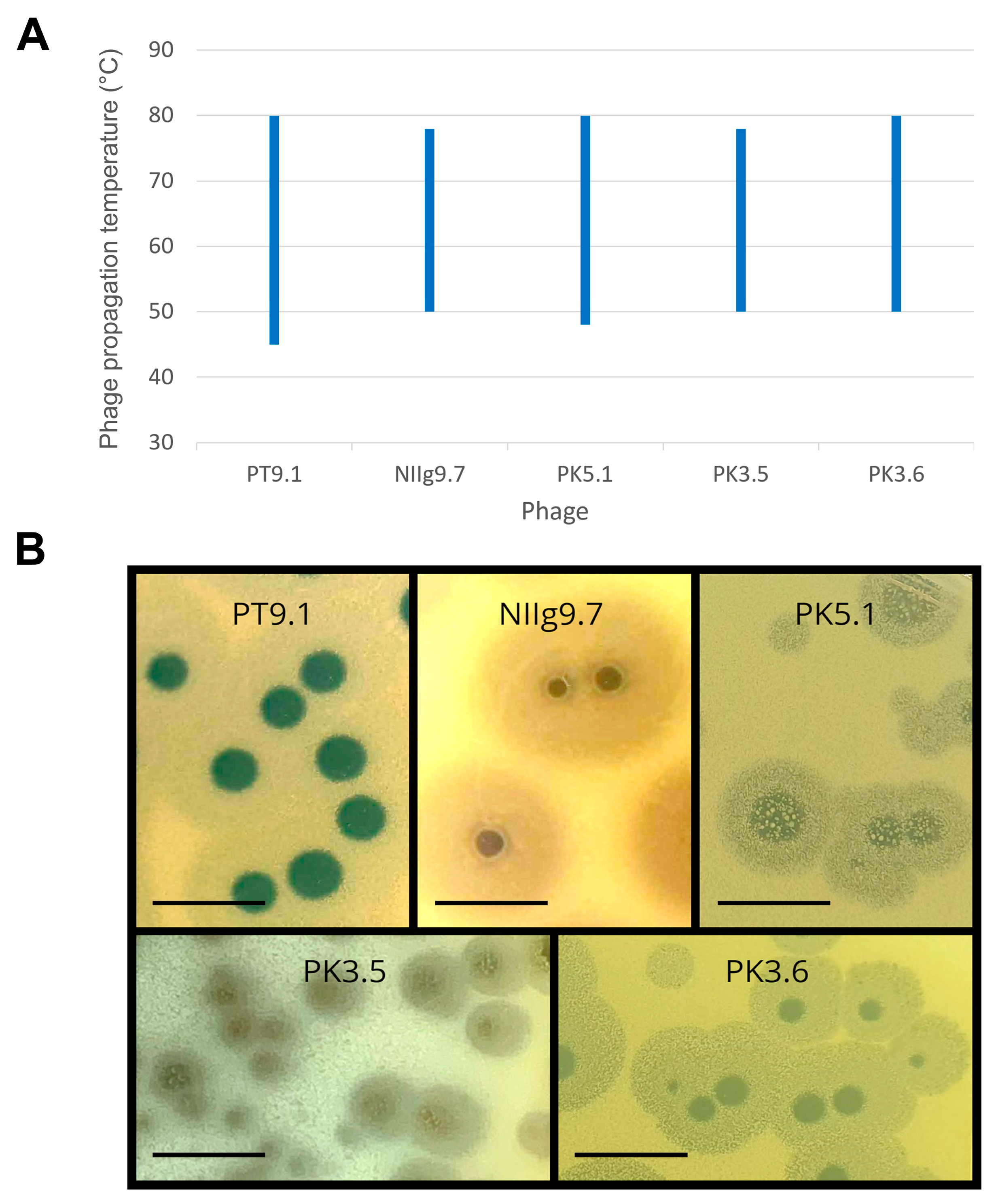



| Phage | Phage Morphology | Plaque Morphology * | |||
|---|---|---|---|---|---|
| Head Diameter (nm) | Tail Length (nm) | Tail Width (nm) | Diameter of a Clear Center (mm) | Diameter of Plaque with a Halo Zone (mm) | |
| PT9.1 | 62.72 ± 2.34 | 143.15 ± 8.47 | 10.68 ± 1.62 | 3.21 ± 0.22 | 9.43 ± 0.73 |
| NIIg9.7 | 63.81 ± 3.95 | 143.16 ± 4.19 | 9.93 ± 1.62 | 3.04 ± 0.31 | 11.42 ± 0.75 |
| PK5.1 | 62.91 ± 3.20 | 145.74 ± 14.88 | 10.10 ± 1.42 | 3.67 ± 0.50 | 7.20 ± 1.23 |
| PK3.5 | 65.18 ± 3.27 | 149.98 ± 15.01 | 10.84 ± 1.92 | 1.45 ± 0.33 | 4.92 ± 0.98 |
| PK3.6 | 66.93 ± 4.24 | 137.32 ± 5.75 | 9.41 ± 1.84 | 3.01 ± 0.51 | 8.42 ± 0.62 |
| Phage | Genome Size (bp) | GC Content (%) | Coding Capacity (%) | N° of ORFs | Best Sequence Alignment * | Identity (%) (Query Coverage (%)) |
|---|---|---|---|---|---|---|
| PT9.1 | 38,373 | 43.9 | 94.2 | 75 | Geobacillus phage vB_GthS_NIIg9.7 | 97.04 (85) |
| NIIg9.7 | 39,016 | 44.4 | 93.9 | 76 | Geobacillus phage vB_GthS_PT9.1 | 97.04 (83) |
| PK5.1 | 38,161 | 43.6 | 92.4 | 64 | Geobacillus phage vB_GthS_PK3.5 | 89.31 (59) |
| PK3.5 | 38,788 | 43.5 | 92.7 | 76 | Geobacillus phage vB_GthS_PK3.6 | 97.60 (84) |
| PK3.6 | 38,405 | 44.8 | 92.0 | 68 | Geobacillus phage vB_GthS_PK3.5 | 97.60 (85) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimoliūnas, E.; Šimoliūnienė, M.; Laskevičiūtė, G.; Kvederavičiūtė, K.; Skapas, M.; Kaupinis, A.; Valius, M.; Meškys, R.; Kuisienė, N. Geobacillus Bacteriophages from Compost Heaps: Representatives of Three New Genera within Thermophilic Siphoviruses. Viruses 2023, 15, 1691. https://doi.org/10.3390/v15081691
Šimoliūnas E, Šimoliūnienė M, Laskevičiūtė G, Kvederavičiūtė K, Skapas M, Kaupinis A, Valius M, Meškys R, Kuisienė N. Geobacillus Bacteriophages from Compost Heaps: Representatives of Three New Genera within Thermophilic Siphoviruses. Viruses. 2023; 15(8):1691. https://doi.org/10.3390/v15081691
Chicago/Turabian StyleŠimoliūnas, Eugenijus, Monika Šimoliūnienė, Gintarė Laskevičiūtė, Kotryna Kvederavičiūtė, Martynas Skapas, Algirdas Kaupinis, Mindaugas Valius, Rolandas Meškys, and Nomeda Kuisienė. 2023. "Geobacillus Bacteriophages from Compost Heaps: Representatives of Three New Genera within Thermophilic Siphoviruses" Viruses 15, no. 8: 1691. https://doi.org/10.3390/v15081691
APA StyleŠimoliūnas, E., Šimoliūnienė, M., Laskevičiūtė, G., Kvederavičiūtė, K., Skapas, M., Kaupinis, A., Valius, M., Meškys, R., & Kuisienė, N. (2023). Geobacillus Bacteriophages from Compost Heaps: Representatives of Three New Genera within Thermophilic Siphoviruses. Viruses, 15(8), 1691. https://doi.org/10.3390/v15081691







