Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany
Abstract
1. Introduction
2. Materials and Methods
2.1. Surveillance
2.2. Wastewater Sampling and Quantification of Viral Load
2.3. Sequencing of SARS-CoV-2 RNA Fragments in Wastewater
2.4. Statistical Analysis
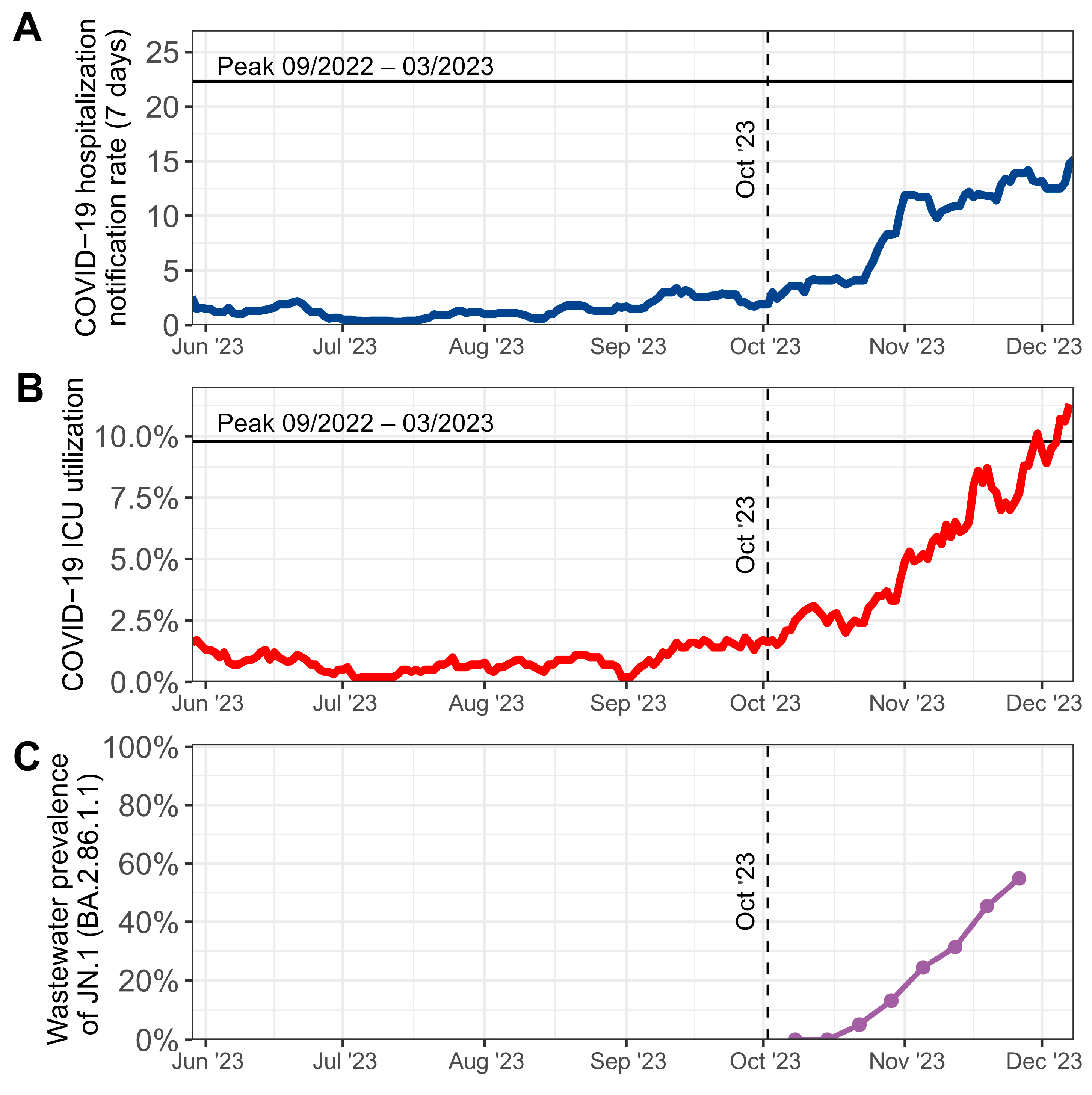
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Zhakparov, D.; Quirin, Y.; Xiao, Y.; Battaglia, N.; Holzer, M.; Bühler, M.; Kistler, W.; Engel, D.; Zumthor, J.P.; Caduff, A.; et al. Sequencing of SARS-CoV-2 RNA Fragments in Wastewater Detects the Spread of New Variants during Major Events. Microorganisms 2023, 11, 2660. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global Initiative on Sharing All Influenza Data—From Vision to Reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef]
- McManus, O.; Christiansen, L.E.; Nauta, M.; Krogsgaard, L.W.; Bahrenscheer, N.S.; Von Kappelgaard, L.; Christiansen, T.; Hansen, M.; Hansen, N.C.; Kähler, J.; et al. Predicting COVID-19 Incidence Using Wastewater Surveillance Data, Denmark, October 2021–June 2022. Emerg. Infect. Dis. 2023, 29, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Gongora, C.; Berg, C.; Rehn, M.; Varg, J.E.; Dillner, L.; Latorre-Margalef, N.; Székely, A.J.; Andersson, E.; Movert, E. Early Detection of the Emerging SARS-CoV-2 BA.2.86 Lineage through Integrated Genomic Surveillance of Wastewater and COVID-19 Cases in Sweden, Weeks 31 to 38 2023. Eurosurveillance 2023, 28, 2300595. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater Sequencing Reveals Early Cryptic SARS-CoV-2 Variant Transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Robert Koch-Institut. COVID-19-Hospitalisierungen in Deutschland. Zenodo 2023, 5519056. [Google Scholar] [CrossRef]
- Robert Koch-Institut. Intensivkapazitäten und COVID-19-Intensivbettenbelegung in Deutschland. Zenodo 2023, 10260666. [Google Scholar] [CrossRef]
- The European Monitoring of Excess Mortality for Public Health Action (EuroMOMO) Network EuroMOMO. Copenhagen: EuroMOMO. Available online: http://www.euromomo.eu (accessed on 21 December 2023).
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Van Boven, M.; Hetebrij, W.A.; Swart, A.; Nagelkerke, E.; Van Der Beek, R.F.; Stouten, S.; Hoogeveen, R.T.; Miura, F.; Kloosterman, A.; Van Der Drift, A.-M.R.; et al. Patterns of SARS-CoV-2 Circulation Revealed by a Nationwide Sewage Surveillance Programme, the Netherlands, August 2020 to February 2022. Eurosurveillance 2023, 28, 2200700. [Google Scholar] [CrossRef] [PubMed]
- Robert Koch-Institut. SARS-CoV-2 Sequenzdaten aus Deutschland. Zenodo 2023, 5139363. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast Evolution of SARS-CoV-2 BA.2.86 to JN.1 under Heavy Immune Pressure. BioRxiv Prepr. 2023, 2023.11.13.566860. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.-H.; Jia, Y.-J.; Zhao, D.-Y.; Fu, X.-L.; Zhang, W.-Q.; Tang, W.; Hu, S.-Q.; Wu, H.; Ge, M.-W.; Du, W.; et al. Clinical Outcomes of the Severe Acute Respiratory Syndrome Coronavirus 2 Omicron and Delta Variant: Systematic Review and Meta-Analysis of 33 Studies Covering 6 037 144 Coronavirus Disease 2019–Positive Patients. Clin. Microbiol. Infect. 2023, 29, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Nextstrain Nextclade Datasets Update 2023-10-26/Github. Available online: https://github.com/nextstrain/nextclade_data (accessed on 4 December 2023).
- O’Toole, Á.; Pybus, O.G.; Abram, M.E.; Kelly, E.J.; Rambaut, A. Pango Lineage Designation and Assignment Using SARS-CoV-2 Spike Gene Nucleotide Sequences. BMC Genom. 2022, 23, 121. [Google Scholar] [CrossRef] [PubMed]
- European Parliament and Council Commission Recommendation (EU) 2021/472 of 17 March 2021 on a Common Approach to Establish a Systematic Surveillance of SARS-CoV-2 and Its Variants in Wastewaters in the EU. Available online: https://eur-lex.europa.eu/eli/reco/2021/472/oj (accessed on 4 December 2023).
- Amman, F.; Markt, R.; Endler, L.; Hupfauf, S.; Agerer, B.; Schedl, A.; Richter, L.; Zechmeister, M.; Bicher, M.; Heiler, G.; et al. Viral Variant-Resolved Wastewater Surveillance of SARS-CoV-2 at National Scale. Nat. Biotechnol. 2022, 40, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Lara, R.; Elsinga, G.; Heijnen, L.; Munnink, B.B.O.; Schapendonk, C.M.E.; Nieuwenhuijse, D.; Kon, M.; Lu, L.; Aarestrup, F.M.; Lycett, S.; et al. Monitoring SARS-CoV-2 Circulation and Diversity through Community Wastewater Sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021, 27, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Wurtz, N.; Boussier, M.; Souville, L.; Penant, G.; Lacoste, A.; Colson, P.; La Scola, B.; Aherfi, S. Simple Wastewater Preparation Protocol Applied to Monitor the Emergence of the Omicron 21L/BA.2 Variant by Genome Sequencing. Viruses 2023, 15, 268. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartel, A.; Grau, J.H.; Bitzegeio, J.; Werber, D.; Linzner, N.; Schumacher, V.; Garske, S.; Liere, K.; Hackenbeck, T.; Rupp, S.I.; et al. Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses 2024, 16, 102. https://doi.org/10.3390/v16010102
Bartel A, Grau JH, Bitzegeio J, Werber D, Linzner N, Schumacher V, Garske S, Liere K, Hackenbeck T, Rupp SI, et al. Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses. 2024; 16(1):102. https://doi.org/10.3390/v16010102
Chicago/Turabian StyleBartel, Alexander, José Horacio Grau, Julia Bitzegeio, Dirk Werber, Nico Linzner, Vera Schumacher, Sonja Garske, Karsten Liere, Thomas Hackenbeck, Sofia Isabell Rupp, and et al. 2024. "Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany" Viruses 16, no. 1: 102. https://doi.org/10.3390/v16010102
APA StyleBartel, A., Grau, J. H., Bitzegeio, J., Werber, D., Linzner, N., Schumacher, V., Garske, S., Liere, K., Hackenbeck, T., Rupp, S. I., Sagebiel, D., Böckelmann, U., & Meixner, M. (2024). Timely Monitoring of SARS-CoV-2 RNA Fragments in Wastewater Shows the Emergence of JN.1 (BA.2.86.1.1, Clade 23I) in Berlin, Germany. Viruses, 16(1), 102. https://doi.org/10.3390/v16010102







