Glycoprotein 5-Derived Peptides Induce a Protective T-Cell Response in Swine against the Porcine Reproductive and Respiratory Syndrome Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptides
2.2. Animal studies
Experimental Design, Animals, and Housing
2.3. Blood Sampling and Isolation of Peripheral Blood Mononuclear Cells
2.4. Cytokine Detection Assay Panel
2.5. Cytometric Analysis
2.6. In Vitro Assays
Plasmid and Cell Lines
2.7. In Vitro Transcription and Virus Recovery
2.8. RNA Extraction, Retro Transcription, Endpoint PCR, and Real-Time PCR Assay for Virus Detection
2.9. Standard Curve for PRRRSV Quantification
2.10. Co-Cultures of Primary Lymphocytes with PK-15 S10-CD163 Cells Infected with PRRSV
2.11. Statistical Analysis
3. Results
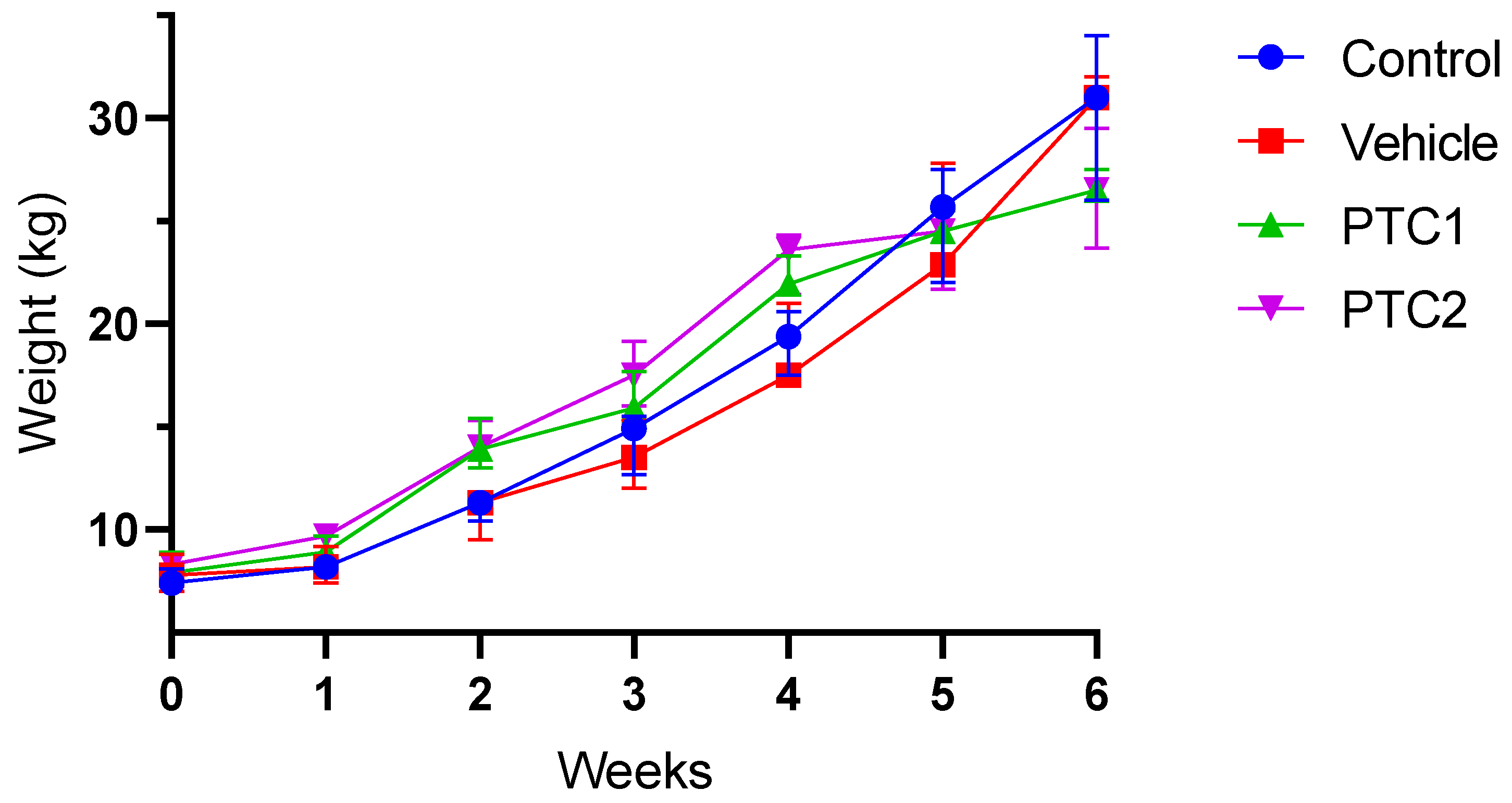
3.1. Weight Gain Was Not Influenced by the Immunization Protocol
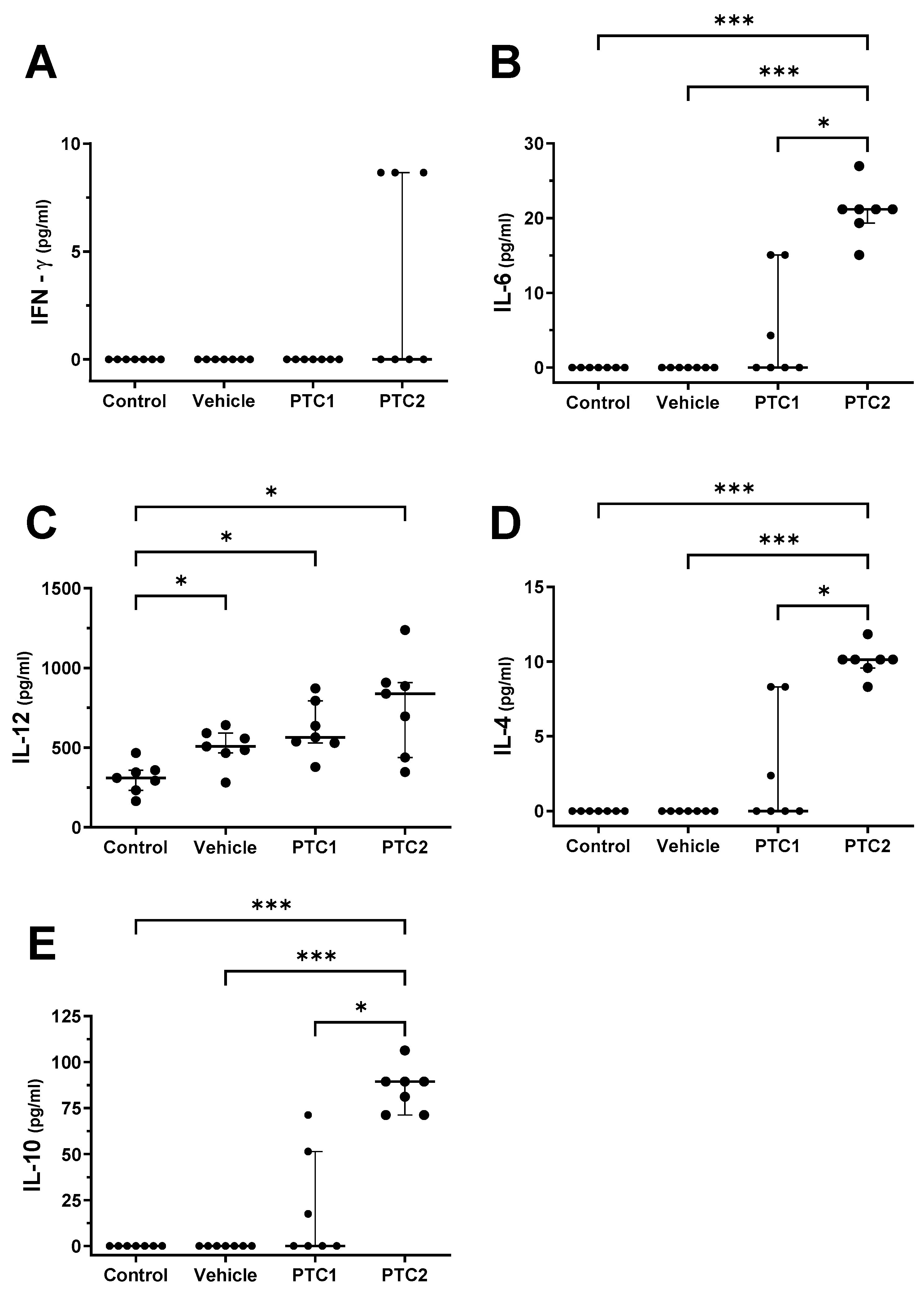
3.2. PTC2 Peptide Increased the Concentration of Proinflammatory Cytokines
3.3. PTC2 Peptide Increased the Serum Concentration of Cytokines Related to the Activation of Cellular Immunity
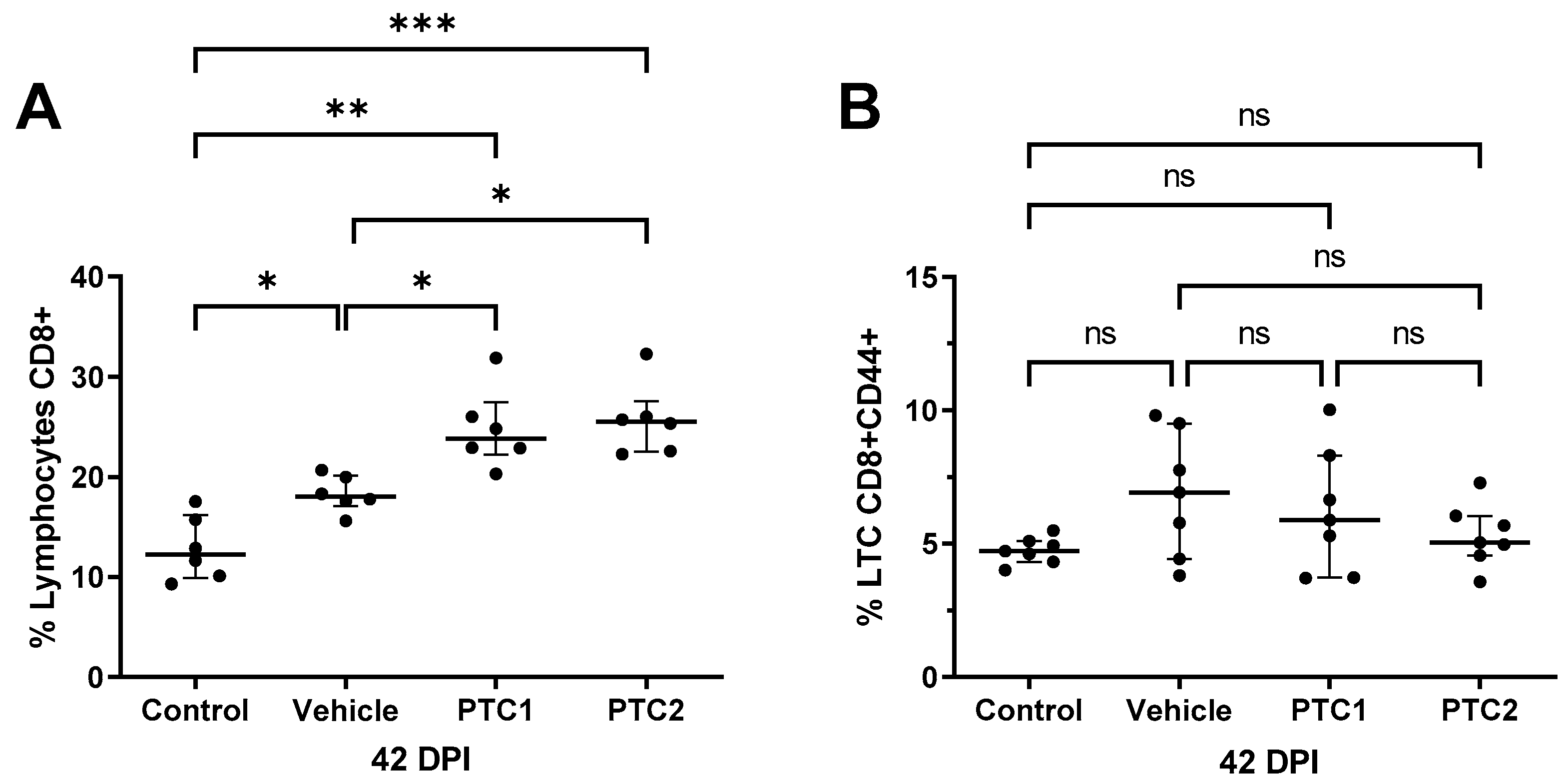
3.4. PTC1 and PTC2 Immunizations Correlated with Increased CD8+ Cell Count in Peripheral Blood
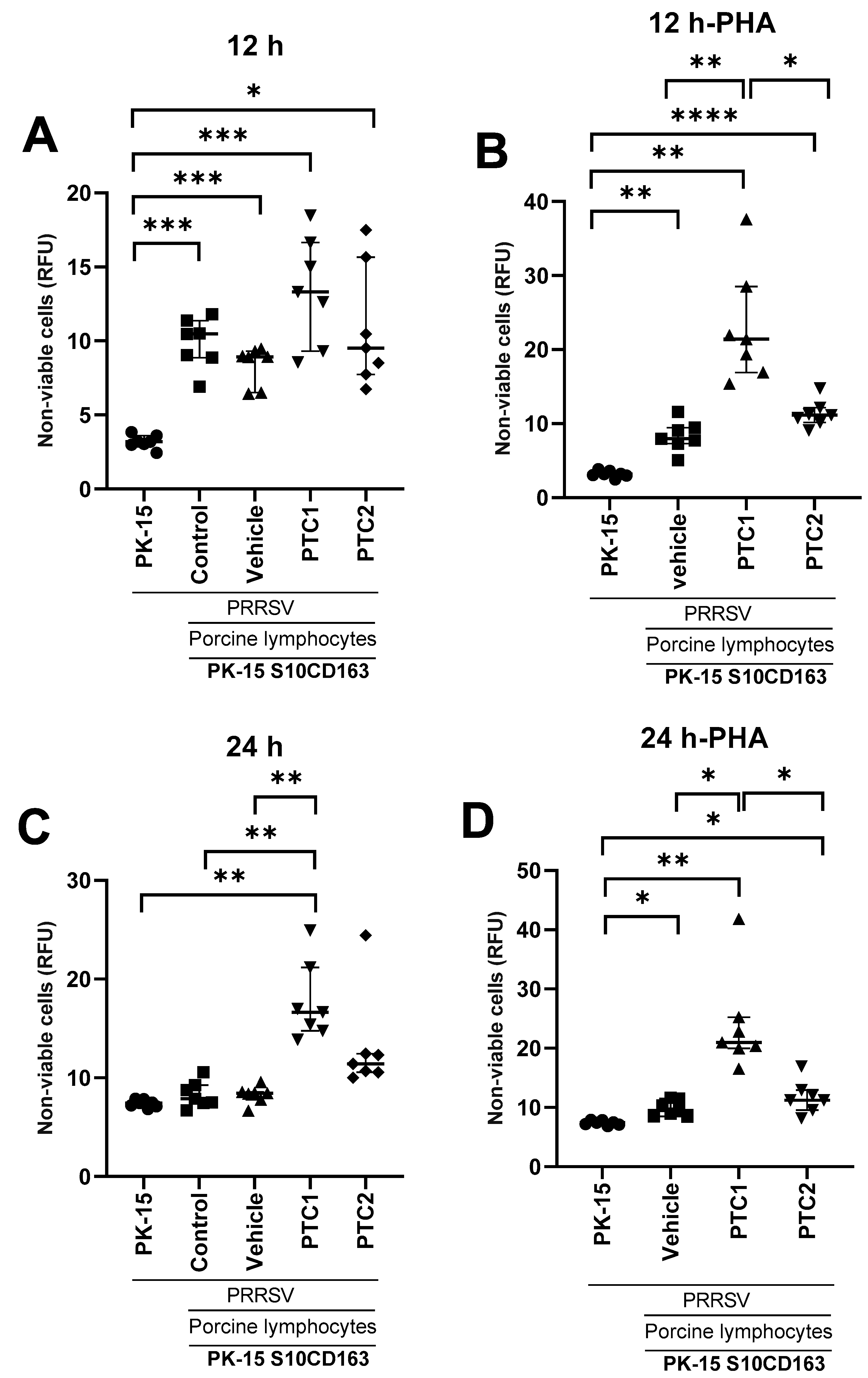
3.5. PTC1 Primed Lymphocytes Recognize and Eliminate PRRSV Infected Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franzo, G.; Barbierato, G.; Pesente, P.; Legnardi, M.; Tucciarone, C.M.; Sandri, G.; Drigo, M. Porcine Reproductive and Respiratory Syndrome (PRRS) Epidemiology in an Integrated Pig Company of Northern Italy: A Multilevel Threat Requiring Multilevel Interventions. Viruses 2021, 13, 2510. [Google Scholar] [CrossRef] [PubMed]
- Jantafong, T.; Sangtong, P.; Saenglub, W.; Mungkundar, C.; Romlamduan, N.; Lekchareonsuk, C.; Lekcharoensuk, P. Genetic Diversity of Porcine Reproductive and Respiratory Syndrome Virus in Thailand and Southeast Asia from 2008 to 2013. Vet. Microbiol. 2015, 176, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yang, H. Porcine Reproductive and Respiratory Syndrome in China. Virus Res. 2010, 154, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Benfield, D.A.; Nelson, E.; Collins, J.E.; Harris, L.; Goyal, S.M.; Robison, D.; Christianson, W.T.; Morrison, R.B.; Gorcyca, D.; Chladek, D. Characterization of Swine Infertility and Respiratory Syndrome (SIRS) Virus (Isolate ATCC VR-2332). J. Vet. Diagn. Investig. 1992, 4, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Meulenberg, J.J.M. The Molecular Biology of Arteriviruses. J. Gen. Virol. 1998, 79, 961–980. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Yoo, D. Engineering the PRRS Virus Genome: Updates and Perspectives. Vet. Microbiol. 2014, 174, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, G.; Yu, L.; Li, L.; Zhang, Y.; Zhou, Y.; Tong, W.; Liu, C.; Gao, F.; Tong, G. Genetic Diversity of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) From 1996 to 2017 in China. Front. Microbiol. 2020, 11, 618. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, S.; Quan, D.; Wang, H.; Huang, J.; Wang, Y.; Ren, T.; Ouyang, K.; Chen, Y.; Huang, W.; et al. Full Genomic Analysis of New Variants of Porcine Reproductive and Respiratory Syndrome Virus Revealed Multiple Recombination Events Between Different Lineages and Sublineages. Front. Vet. Sci. 2020, 7, 603. [Google Scholar] [CrossRef]
- Fang, Y.; Snijder, E.J. The PRRSV Replicase: Exploring the Multifunctionality of an Intriguing Set of Nonstructural Proteins. Virus Res. 2010, 154, 61–76. [Google Scholar] [CrossRef]
- Kappes, M.A.; Faaberg, K.S. PRRSV Structure, Replication and Recombination: Origin of Phenotype and Genotype Diversity. Virology 2015, 479, 475–486. [Google Scholar] [CrossRef]
- Swine Health Information Center. Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus; Swine Health Information Center: Ames, IA, USA, 2016. [Google Scholar]
- Sun, W.; Wu, W.; Jiang, N.; Ge, X.; Zhang, Y.; Han, J.; Guo, X.; Zhou, L.; Yang, H. Highly Pathogenic PRRSV-Infected Alveolar Macrophages Impair the Function of Pulmonary Microvascular Endothelial Cells. Viruses 2022, 14, 452. [Google Scholar] [CrossRef] [PubMed]
- Assavacheep, P.; Thanawongnuwech, R. Porcine Respiratory Disease Complex: Dynamics of Polymicrobial Infections and Management Strategies after the Introduction of the African Swine Fever. Front. Vet. Sci. 2022, 9, 1048861. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, S.; Halbur, P.; Thacker, E. Porcine Respiratory Disease Complex. In Polymicrobial Diseases; Brogden, K.A., Guthmiller, J.M.E., Eds.; ASM Press: Washington, DC, USA, 2002. [Google Scholar]
- Neumann, E.J.; Kliebenstein, J.B.; Johnson, C.D.; Mabry, J.W.; Bush, E.J.; Seitzinger, A.H.; Green, A.L.; Zimmerman, J.J. Assessment of the Economic Impact of Porcine Reproductive and Respiratory Syndrome on Swine Production in the United States. J. Am. Vet. Med. Assoc. 2005, 227, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Z.; Li, H.; Yang, S.; Ren, F.; Bian, T.; Sun, L.; Zhou, B.; Zhou, L.; Qu, X. The Economic Impact of Porcine Reproductive and Respiratory Syndrome Outbreak in Four Chinese Farms: Based on Cost and Revenue Analysis. Front. Vet. Sci. 2022, 9, 1024720. [Google Scholar] [CrossRef] [PubMed]
- Tait-Burkard, C.; Doeschl-Wilson, A.; McGrew, M.J.; Archibald, A.L.; Sang, H.M.; Houston, R.D.; Whitelaw, C.B.; Watson, M. Livestock 2.0—Genome Editing for Fitter, Healthier, and More Productive Farmed Animals. Genome Biol. 2018, 19, 204. [Google Scholar] [CrossRef] [PubMed]
- Chase-Topping, M.; Xie, J.; Pooley, C.; Trus, I.; Bonckaert, C.; Rediger, K.; Bailey, R.I.; Brown, H.; Bitsouni, V.; Barrio, M.B.; et al. New Insights about Vaccine Effectiveness: Impact of Attenuated PRRS-Strain Vaccination on Heterologous Strain Transmission. Vaccine 2020, 38, 3050–3061. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Z.; Chen, K.; Qian, J.; Hu, Y.; Fang, S.; Sun, Z.; Zhang, C.; Huang, L.; Zhang, J.; et al. Efficacy of the Synergy Between Live-Attenuated and Inactivated PRRSV Vaccines Against a NADC30-Like Strain of Porcine Reproductive and Respiratory Syndrome Virus in 4-Week Piglets. Front. Vet. Sci. 2022, 9, 812040. [Google Scholar] [CrossRef]
- Brinton, M.A.; Snijder, E.J. Arteriviruses. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, MA, USA, 2008; pp. 176–186. ISBN 978-0-12-374410-4. [Google Scholar]
- Kvisgaard, L.K.; Kristensen, C.S.; Ryt-Hansen, P.; Pedersen, K.; Stadejek, T.; Trebbien, R.; Andresen, L.O.; Larsen, L.E. A Recombination between Two Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-1) Vaccine Strains Has Caused Severe Outbreaks in Danish Pigs. Transbound. Emerg. Dis. 2020, 67, 1786–1796. [Google Scholar] [CrossRef]
- Geldhof, M.F.; Vanhee, M.; Van Breedam, W.; Van Doorsselaere, J.; Karniychuk, U.U.; Nauwynck, H.J. Comparison of the Efficacy of Autogenous Inactivated Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Vaccines with That of Commercial Vaccines against Homologous and Heterologous Challenges. BMC Vet. Res. 2012, 8, 182. [Google Scholar] [CrossRef]
- Lowe, J.F.; Zuckermann, F.A.; Firkins, L.D.; Schnitzlein, W.M.; Goldberg, T.L. Immunologic Responses and Reproductive Outcomes Following Exposure to Wild-Type or Attenuated Porcine Reproductive and Respiratory Syndrome Virus in Swine under Field Conditions. J. Am. Vet. Med. Assoc. 2006, 228, 1082–1088. [Google Scholar] [CrossRef]
- Thanawongnuwech, R.; Suradhat, S. Taming PRRSV: Revisiting the Control Strategies and Vaccine Design. Virus Res. 2010, 154, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Galliher-Beckley, A.; Huang, H.; Sun, X.; Shi, J. Peptide Nanofiber Hydrogel Adjuvanted Live Virus Vaccine Enhances Cross-Protective Immunity to Porcine Reproductive and Respiratory Syndrome Virus. Vaccine 2013, 31, 4508–4515. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Common Ingredients in U.S. Licensed Vaccines; U.S. Food and Drug Administration: Rockville, MD, USA, 2019.
- Rezaei, M.; Nazari, M. New Generation Vaccines for COVID-19 Based on Peptide, Viral Vector, Artificial Antigen Presenting Cell, DNA or MRNA. Avicenna J. Med. Biotechnol. 2022, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Wu, C.; Gu, G.; Sun, W.; Zhang, Y.-J.; Zhou, E.-M. Improved Vaccine against PRRSV: Current Progress and Future Perspective. Front. Microbiol. 2017, 8, 1635. [Google Scholar] [CrossRef] [PubMed]
- Renukaradhya, G.J.; Meng, X.J.; Calvert, J.G.; Roof, M.; Lager, K.M. Inactivated and Subunit Vaccines against Porcine Reproductive and Respiratory Syndrome: Current Status and Future Direction. Vaccine 2015, 33, 3065–3072. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Murtaugh, M.P. Dissociation of Porcine Reproductive and Respiratory Syndrome Virus Neutralization from Antibodies Specific to Major Envelope Protein Surface Epitopes. Virology 2012, 433, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Petrini, S.; Ramadori, G.; Villa, R.; Borghetti, P.; de Angelis, E.; Cantoni, A.; Corradi, A.; Amici, A.; Ferrari, M. Evaluation of Different DNA Vaccines against Porcine Reproductive and Respiratory Syndrome (PRRS) in Pigs. Vaccines 2013, 1, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Vanhee, M.; Van Breedam, W.; Costers, S.; Geldhof, M.; Noppe, Y.; Nauwynck, H. Characterization of Antigenic Regions in the Porcine Reproductive and Respiratory Syndrome Virus by the Use of Peptide-Specific Serum Antibodies. Vaccine 2011, 29, 4794–4804. [Google Scholar] [CrossRef]
- Zhou, C.; Lu, R.; Lin, G.; Yao, Z. The Latest Developments in Synthetic Peptides with Immunoregulatory Activities. Peptides 2011, 32, 408–414. [Google Scholar] [CrossRef]
- Tondini, E.; Reintjens, N.R.M.; Castello, G.; Arakelian, T.; Isendoorn, M.; Camps, M.; Vree, J.; van der Marel, G.A.; Filippov, D.V.; Codee, J.D.C.; et al. Lipid A Analog CRX-527 Conjugated to Synthetic Peptides Enhances Vaccination Efficacy and Tumor Control. NPJ Vaccines 2022, 7, 64. [Google Scholar] [CrossRef]
- Cox, R. Correlates of Protection to Influenza Virus, Where Do We Go from Here? Hum. Vaccines Immunother. 2013, 9, 405–408. [Google Scholar] [CrossRef] [PubMed]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of Protection against SARS-CoV-2 in Rhesus Macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Rimmelzwaan, G.F.; Fouchier, R.A.M.; Osterhaus, A.D.M.E. Influenza Virus-Specific Cytotoxic T Lymphocytes: A Correlate of Protection and a Basis for Vaccine Development. Curr. Opin. Biotechnol. 2007, 18, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.M.; Tian, D.; Heffron, C.L.; Subramaniam, S.; Opriessnig, T.; Foss, D.L.; Calvert, J.G.; Meng, X.-J. Cytotoxic T Lymphocyte Epitopes Identified from a Contemporary Strain of Porcine Reproductive and Respiratory Syndrome Virus Enhance CD4+CD8+ T, CD8+ T, and Γδ T Cell Responses. Virology 2019, 538, 35–44. [Google Scholar] [CrossRef]
- Welner, S.; Nielsen, M.; Rasmussen, M.; Buus, S.; Jungersen, G.; Larsen, L.E. Prediction and in Vitro Verification of Potential CTL Epitopes Conserved among PRRSV-2 Strains. Immunogenetics 2017, 69, 689–702. [Google Scholar] [CrossRef]
- Vashisht, K.; Goldberg, T.L.; Husmann, R.J.; Schnitzlein, W.; Zuckermann, F.A. Identification of Immunodominant T-Cell Epitopes Present in Glycoprotein 5 of the North American Genotype of Porcine Reproductive and Respiratory Syndrome Virus. Vaccine 2008, 26, 4747–4753. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Ansari, I.H.; Kwon, B.; Osorio, F.A.; Pattnaik, A.K. Influence of N-Linked Glycosylation of Porcine Reproductive and Respiratory Syndrome Virus GP5 on Virus Infectivity, Antigenicity, and Ability to Induce Neutralizing Antibodies. J. Virol. 2006, 80, 3994–4004. [Google Scholar] [CrossRef]
- Woźniakowski, G.; Samorek-Salamonowicz, E.; Kozdruń, W. Quantitative Analysis of Waterfowl Parvoviruses in Geese and Muscovy Ducks by Real-Time Polymerase Chain Reaction: Correlation between Age, Clinical Symptoms and DNA Copy Number of Waterfowl Parvoviruses. BMC Vet. Res. 2012, 8, 29. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Hoskin, D.W.; Power Coombs, M.R. Anticancer Activities of Natural and Synthetic Peptides. Adv. Exp. Med. Biol. 2019, 1117, 131–147. [Google Scholar] [CrossRef]
- He, P.; Zou, Y.; Hu, Z. Advances in Aluminum Hydroxide-Based Adjuvant Research and Its Mechanism. Hum. Vaccines Immunother. 2015, 11, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, J.; Bi, Y.; Yang, L.; Meng, S.; Zhou, Y.; Jia, X.; Meng, S.; Sun, L.; Liu, W. Synthetic B- and T-Cell Epitope Peptides of Porcine Reproductive and Respiratory Syndrome Virus with Gp96 as Adjuvant Induced Humoral and Cell-Mediated Immunity. Vaccine 2013, 31, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Marsman, C.; Jorritsma, T.; Ten Brinke, A.; van Ham, S.M. Flow Cytometric Methods for the Detection of Intracellular Signaling Proteins and Transcription Factors Reveal Heterogeneity in Differentiating Human B Cell Subsets. Cells 2020, 9, 2633. [Google Scholar] [CrossRef] [PubMed]
- Díaz, I.; Pujols, J.; Ganges, L.; Gimeno, M.; Darwich, L.; Domingo, M.; Mateu, E. In Silico Prediction and Ex Vivo Evaluation of Potential T-Cell Epitopes in Glycoproteins 4 and 5 and Nucleocapsid Protein of Genotype-I (European) of Porcine Reproductive and Respiratory Syndrome Virus. Vaccine 2009, 27, 5603–5611. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, C.; Takamatsu, H.; Âoni, C.A.; Audonnet, J.-C.; Fischer, L.; Lefe Ávre, F.; Charley, B. Enhanced Protective Response and Immuno-Adjuvant Effects of Porcine GM-CSF on DNA Vaccination of Pigs against Aujeszky’s Disease Virus. Vet. Immunol. Immunopathol. 1999, 70, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lee, A.; Grigoryan, L.; Arunachalam, P.S.; Scott, M.K.D.; Trisal, M.; Wimmers, F.; Sanyal, M.; Weidenbacher, P.A.; Feng, Y.; et al. Mechanisms of Innate and Adaptive Immunity to the Pfizer-BioNTech BNT162b2 Vaccine. Nat. Immunol. 2022, 23, 543–555. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Scott, M.K.D.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L. Systems Vaccinology of the BNT162b2 MRNA Vaccine in Humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef]
- Mountford, A.P.; Coulson, P.S.; Cheever, A.W.; Sher, A.; Wilson, R.A.; Wynn, T.A. Interleukin-12 Can Directly Induce T-Helper 1 Responses in Interferon-c (IFN-c) Receptor-Deficient Mice, but Requires IFN-c Signalling to Downregulate T-Helper 2 Responses. Immunology 1999, 97, 588–594. [Google Scholar] [CrossRef]
- Rincón, M.; Anguita, J.; Nakamura, T.; Fikrig, E.; Flavell, R.A. Interleukin (IL)-6 Directs the Differentiation of IL-4-Producing CD4 T Cells. J. Exp. Med. 1997, 185, 461–470. [Google Scholar] [CrossRef]
- Heijink, I.H.; Vellenga, E.; Borger, P.; Postma, D.S.; De Monchy, J.G.R.; Kauffman, H.F. Interleukin-6 Promotes the Production of Interleukin-4 and Interleukin-5 by Interleukin-2-Dependent and-Independent Mechanisms in Freshly Isolated Human T Cells. Immunology 2002, 107, 316–324. [Google Scholar] [CrossRef]
- Chung, N.H.; Chen, Y.C.; Yang, S.J.; Lin, Y.C.; Dou, H.Y.; Hui-Ching Wang, L.; Liao, C.L.; Chow, Y.H. Induction of Th1 and Th2 in the Protection against SARS-CoV-2 through Mucosal Delivery of an Adenovirus Vaccine Expressing an Engineered Spike Protein. Vaccine 2022, 40, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Xia, Q.; Zhou, J.; Liu, H.; Chen, Y.; Liu, Y.; Ding, P.; Qi, Y.; Wang, A. Identification of Potential SLA-I-Restricted CTL Epitopes within the M Protein of Porcine Reproductive and Respiratory Syndrome Virus. Vet. Microbiol. 2021, 259, 109131. [Google Scholar] [CrossRef] [PubMed]
- Mokhtar, H.; Pedrera, M.; Frossard, J.P.; Biffar, L.; Hammer, S.E.; Kvisgaard, L.K.; Larsen, L.E.; Stewart, G.R.; Somavarapu, S.; Steinbach, F.; et al. The Non-Structural Protein 5 and Matrix Protein Are Antigenic Targets of T Cell Immunity to Genotype 1 Porcine Reproductive and Respiratory Syndrome Viruses. Front. Immunol. 2016, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lin, Y.; Bai, Y.; Tong, T.; Wang, Q.; Liu, N.; Liu, G.; Xiao, Y.; Yang, T.; Bu, Z. Identification of CD8+ Cytotoxic T Lymphocyte Epitopes from Porcine Reproductive and Respiratory Syndrome Virus Matrix Protein in BALB/c Mice. Virol. J. 2011, 8, 263. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-C.; Lin, H.-H.; Lin, C.-H.; Chung, W.-B. Identification of Cytotoxic T Lymphocyte Epitopes on Swine Viruses: Multi-Epitope Design for Universal T Cell Vaccine. PLoS ONE 2013, 8, e84443. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhang, N.; Wei, X.; Jiang, Y.; Chen, R.; Li, Q.; Liang, R.; Zhang, L.; Ma, L.; Xia, C. Illumination of PRRSV Cytotoxic T Lymphocyte Epitopes by the Three-Dimensional Structure and Peptidome of Swine Lymphocyte Antigen Class I (SLA-I). Front. Immunol. 2020, 10, 2995. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Luo, M.; Wei, Y. Alhydrogel—Sodium Chloride Compound Immunologic Adjuvant, Preparation Method and Use Thereof. United States Patent 5861366, 22 December 2020. [Google Scholar]
- Sheikh, N.A.; Rajananthanan, P.; Attard, G.S.; Morrow, W.J.W. Generation of Antigen Specific CD8+ Cytotoxic T Cells Following Immunization with Soluble Protein Formulated with Novel Glycoside Adjuvants. Vaccine 1999, 17, 2974–2982. [Google Scholar] [CrossRef]
- Franzoni, G.; Kurkure, N.V.; Edgar, D.S.; Everett, H.E.; Gerner, W.; Bodman-Smith, K.B.; Crooke, H.R.; Grahama, S.P. Assessment of the Phenotype and Functionality of Porcine Cd8 t Cell Responses Following Vaccination with Live Attenuated Classical Swine Fever Virus (CSFV) and Virulent CSFV Challenge. Clin. Vaccine Immunol. 2013, 20, 1604–1616. [Google Scholar] [CrossRef]
- Mendieta, I.; Nuñez-Anita, R.E.; Pérez-Sánchez, G.; Pavón, L.; Rodríguez-Cruz, A.; García-Alcocer, G.; Berumen, L.C. Effect of A549 Neuroendocrine Differentiation on Cytotoxic Immune Response. Endocr. Connect. 2018, 7, 791–802. [Google Scholar] [CrossRef]
- Siedlik, J.A.; Deckert, J.A.; Benedict, S.H.; Bhatta, A.; Dunbar, A.J.; Vardiman, J.P.; Gallagher, P.M. T Cell Activation and Proliferation Following Acute Exercise in Human Subjects Is Altered by Storage Conditions and Mitogen Selection. J. Immunol. Methods 2017, 446, 7–14. [Google Scholar] [CrossRef]
- Chilson, O.P.; Boylston, A.W.; Crumpton, M.J. Phaseolus Vulgaris Phytohaemagglutinin (PHA) Binds to the Human T Lymphocyte Antigen Receptor. EMBO J. 1984, 3, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Movafagh, A.; Heydary, H.; Mortazavi-Tabatabaei, S.A.; Azargashb, E. The Significance Application of Indigenous Phytohemagglutinin (PHA) Mitogen on Metaphase and Cell Culture Procedure. Iran. J. Pharm. Res. IJPR 2011, 10, 895–903. [Google Scholar] [PubMed]
- Li, Y.; Díaz, I.; Martín-Valls, G.; Beyersdorf, N.; Mateu, E. Systemic CD4 Cytotoxic T Cells Improve Protection against PRRSV-1 Transplacental Infection. Front. Immunol. 2023, 13, 1020227. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calderon-Rico, F.; Bravo-Patiño, A.; Mendieta, I.; Perez-Duran, F.; Zamora-Aviles, A.G.; Franco-Correa, L.E.; Ortega-Flores, R.; Hernandez-Morales, I.; Nuñez-Anita, R.E. Glycoprotein 5-Derived Peptides Induce a Protective T-Cell Response in Swine against the Porcine Reproductive and Respiratory Syndrome Virus. Viruses 2024, 16, 14. https://doi.org/10.3390/v16010014
Calderon-Rico F, Bravo-Patiño A, Mendieta I, Perez-Duran F, Zamora-Aviles AG, Franco-Correa LE, Ortega-Flores R, Hernandez-Morales I, Nuñez-Anita RE. Glycoprotein 5-Derived Peptides Induce a Protective T-Cell Response in Swine against the Porcine Reproductive and Respiratory Syndrome Virus. Viruses. 2024; 16(1):14. https://doi.org/10.3390/v16010014
Chicago/Turabian StyleCalderon-Rico, Fernando, Alejandro Bravo-Patiño, Irasema Mendieta, Francisco Perez-Duran, Alicia Gabriela Zamora-Aviles, Luis Enrique Franco-Correa, Roberto Ortega-Flores, Ilane Hernandez-Morales, and Rosa Elvira Nuñez-Anita. 2024. "Glycoprotein 5-Derived Peptides Induce a Protective T-Cell Response in Swine against the Porcine Reproductive and Respiratory Syndrome Virus" Viruses 16, no. 1: 14. https://doi.org/10.3390/v16010014





