Diagnosis of Imported Dengue and Zika Virus Infections in Italy from November 2015 to November 2022: Laboratory Surveillance Data from a National Reference Laboratory
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Direct Virus Detection Assays
2.2.1. RNA Extraction and Real Time RT-PCRs
2.2.2. Dengue NS1 Antigen Detection
2.3. Serological Assays
2.3.1. Anti-DENV IgM ELISA Systems
2.3.2. Anti-ZIKV IgM ELISA Systems
2.3.3. Plaque Reduction Neutralization Test (PRNT)
2.4. Analysis of Results and Statistics
3. Results
3.1. Cases Clasification and Laboraotory Tests Results
3.1.1. Cases Classification
3.1.2. Demographical, Clinical and Epidemiological Data of Confirmed and Probable Cases
3.1.3. Laboratory Tests Results
3.1.4. Cross-Reactivity between DENV and ZIKV
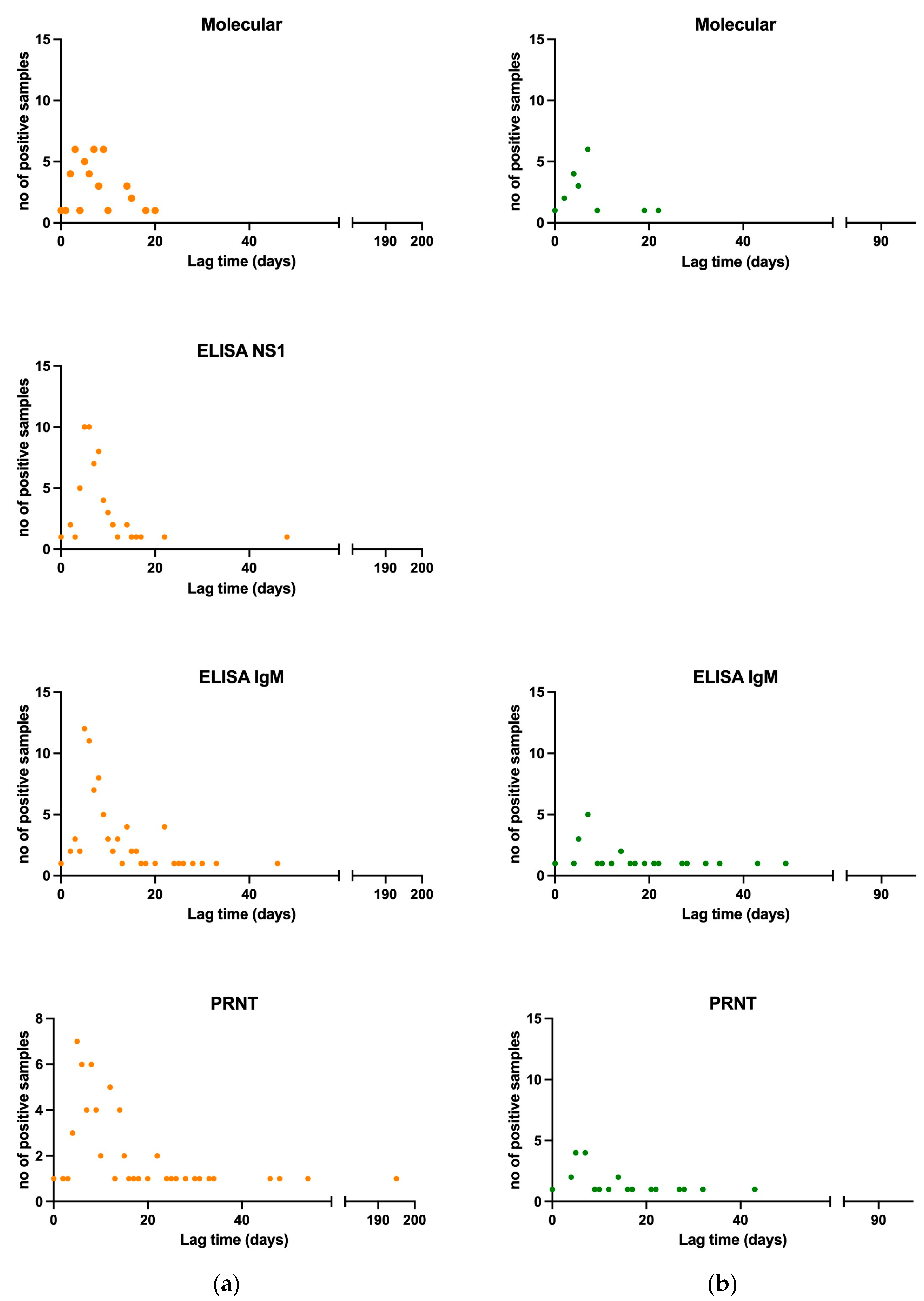
3.1.5. Viral and Antibody Kinetics of Confirmed and Probable Cases
3.1.6. Diagnostic Performance Assessment of Commercially Available ELISA Systems
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wellekens, K.; Betrains, A.; De Munter, P.; Peetermans, W. Dengue: Current State One Year before WHO 2010–2020 Goals. Acta Clin. Belg. 2022, 77, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Beer, M.; Blitvich, B.J.; Bukh, J.; De Lamballerie, X.; Drexler, J.F.; Imrie, A.; Kapoor, A.; Karganova, G.G.; Lemey, P.; et al. Renaming of the Genus Flavivirus to Orthoflavivirus and Extension of Binomial Species Names within the Family Flaviviridae. Arch. Virol. 2023, 168, 224. [Google Scholar] [CrossRef] [PubMed]
- Sittikul, P.; Sriburin, P.; Rattanamahaphoom, J.; Limkittikul, K.; Sirivichayakul, C.; Chatchen, S. Combining Immunoassays to Identify Zika Virus Infection in Dengue-Endemic Areas. Trop. Med. Infect. Dis. 2022, 7, 254. [Google Scholar] [CrossRef] [PubMed]
- van Leur, S.W.; Heunis, T.; Munnur, D.; Sanyal, S. Pathogenesis and Virulence of Flavivirus Infections. Virulence 2021, 12, 2814–2838. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Recombinant Protein–Based Dengue Vaccines. Front. Immunol. 2018, 9, 1919. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, L.T.C.; Tura, B.; Santos, M. Systematic Review of Dengue Vaccine Efficacy. BMC Infect. Dis. 2019, 19, 750. [Google Scholar] [CrossRef]
- Sharma, V.; Sharma, M.; Dhull, D.; Sharma, Y.; Kaushik, S.; Kaushik, S. Zika Virus: An Emerging Challenge to Public Health Worldwide. Can. J. Microbiol. 2020, 66, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.; Adams, L.E.; Durbin, A.P.; Muñoz-Jordán, J.L.; Poehling, K.A.; Sánchez-González, L.M.; Volkman, H.R.; Paz-Bailey, G. Dengue: A Growing Problem With New Interventions. Pediatrics 2022, 149, e2021055522. [Google Scholar] [CrossRef]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef]
- Pang, T.; Mak, T.K.; Gubler, D.J. Prevention and Control of Dengue—The Light at the End of the Tunnel. Lancet Infect. Dis. 2017, 17, e79–e87. [Google Scholar] [CrossRef]
- Schaffner, F.; Mathis, A. Dengue and Dengue Vectors in the WHO European Region: Past, Present, and Scenarios for the Future. Lancet Infect. Dis. 2014, 14, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Benelli, G.; Wilke, A.B.B.; Beier, J.C. Aedes Albopictus (Asian Tiger Mosquito). Trends Parasitol. 2020, 36, 942–943. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Zika Virus; World Health Organisation: Geneva, Switzerland, 2022. [Google Scholar]
- Giron, S.; Franke, F.; Decoppet, A.; Cadiou, B.; Travaglini, T.; Thirion, L.; Durand, G.; Jeannin, C.; L’Ambert, G.; Grard, G.; et al. Vector-Borne Transmission of Zika Virus in Europe, Southern France, August 2019. Eurosurveillance 2019, 24, 1900655. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Dengue and Severe Dengue; World Health Organisation: Geneva, Switzerland, 2023. [Google Scholar]
- Hassan, M.; Ali, S.; Saleem, M.; Sanaullah, M.; Fahad, L.G.; Kim, J.Y.; Alquhayz, H.; Tahir, S.F. Diagnosis of Dengue Virus Infection Using Spectroscopic Images and Deep Learning. PeerJ Comput. Sci. 2022, 8, e985. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Autochthonous Vectorial Transmission of Dengue Virus in Mainland EU/EEA, 2010–Present; European Center for Disease Prevention and Control: Solna, Sweden, 2023. [Google Scholar]
- Cochet, A.; Calba, C.; Jourdain, F.; Grard, G.; Durand, G.A.; Guinard, A.; Investigation team; Noël, H.; Paty, M.-C.; Franke, F. Autochthonous Dengue in Mainland France, 2022: Geographical Extension and Incidence Increase. Eurosurveillance 2022, 27, 2200818. [Google Scholar] [CrossRef]
- Lazzarini, L.; Barzon, L.; Foglia, F.; Manfrin, V.; Pacenti, M.; Pavan, G.; Rassu, M.; Capelli, G.; Montarsi, F.; Martini, S.; et al. First Autochthonous Dengue Outbreak in Italy, August 2020. Eurosurveillance 2020, 25, 2001606. [Google Scholar] [CrossRef]
- Cassaniti, I.; Ferrari, G.; Senatore, S.; Rossetti, E.; Defilippo, F.; Maffeo, M.; Vezzosi, L.; Campanini, G.; Sarasini, A.; Paolucci, S.; et al. Preliminary Results on an Autochthonous Dengue Outbreak in Lombardy Region, Italy, August 2023. Eurosurveillance 2023, 28, 2300471. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Communicable Disease Threats Report, 23–29 July 2023, Week 30; European Center for Disease Prevention and Control: Solna, Sweden, 2023. [Google Scholar]
- Riccardo, F.; Bella, A.; Del Manso, M. Sistema Nazionale Di Sorveglianza Delle Arbovirosi: I Bollettini Periodici 2023. Available online: https://www.epicentro.iss.it/arbovirosi/dashboard (accessed on 19 December 2023).
- Pati, I.; Pisani, G.; Riccardo, F.; Venturi, G.; De Angelis, V. DENV Outbreak in Italy: The Impact on the National Transfusion Network: DENV Outbreak in Italy: The Impact on the National Transfusion Network. Blood Transfus. 2023; ahead of print. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Clinical Presentation; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2023. [Google Scholar]
- Adams, L.E.; Waterman, S.; Paz-Bailey, G. Vaccination for Dengue Prevention. JAMA 2022, 327, 817. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Symptoms; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- Plourde, A.R.; Bloch, E.M. A Literature Review of Zika Virus. Emerg. Infect. Dis. 2016, 22, 1185–1192. [Google Scholar] [CrossRef]
- Thwaites, G.E.; Day, N.P.J. Approach to Fever in the Returning Traveler. N. Engl. J. Med. 2017, 376, 548–560. [Google Scholar] [CrossRef]
- Chan, K.R.; Ismail, A.A.; Thergarajan, G.; Raju, C.S.; Yam, H.C.; Rishya, M.; Sekaran, S.D. Serological Cross-Reactivity among Common Flaviviruses. Front. Cell. Infect. Microbiol. 2022, 12, 975398. [Google Scholar] [CrossRef] [PubMed]
- Songjaeng, A.; Thiemmeca, S.; Mairiang, D.; Punyadee, N.; Kongmanas, K.; Hansuealueang, P.; Tangthawornchaikul, N.; Duangchinda, T.; Mongkolsapaya, J.; Sriruksa, K.; et al. Development of a Singleplex Real-Time Reverse Transcriptase PCR Assay for Pan-Dengue Virus Detection and Quantification. Viruses 2022, 14, 1271. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Factsheet about Dengue; European Center for Disease Prevention and Control: Stockholm, Sweden, 2023. [Google Scholar]
- European Centre for Disease Prevention and Control. Factsheet about Zika Virus Disease; European Center for Disease Prevention and Control: Stockholm, Sweden, 2021. [Google Scholar]
- Cordeiro, M.T. Postnatal Imaging Findings of Congenital Zika Syndrome: The Story of a Disease That Is Still Being Written. Top. Magn. Reson. Imaging 2019, 28, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.-C.; Huang, Y.-Y.; Shu, P.-Y.; Chang, S.-F.; Hsieh, P.-S.; Wey, J.-J.; Tsai, M.-H.; Ben, R.-J.; Hsu, Y.-M.; Fang, Y.-C.; et al. Development of an Enzyme-Linked Immunosorbent Assay for Rapid Detection of Dengue Virus (DENV) NS1 and Differentiation of DENV Serotypes during Early Infection. J. Clin. Microbiol. 2019, 57, e00221-19. [Google Scholar] [CrossRef]
- Narayan, R.; Raja, S.; Kumar, S.; Sambasivam, M.; Jagadeesan, R.; Arunagiri, K.; Krishnasamy, K.; Palani, G. A Novel Indirect ELISA for Diagnosis of Dengue Fever. Indian J. Med. Res. 2016, 144, 128. [Google Scholar] [CrossRef] [PubMed]
- Vairo, F.; Di Pietrantonj, C.; Pasqualini, C.; Mammone, A.; Lanini, S.; Nicastri, E.; Castilletti, C.; Ferraro, F.; Di Bari, V.; Puro, V.; et al. The Surveillance of Chikungunya Virus in a Temperate Climate: Challenges and Possible Solutions from the Experience of Lazio Region, Italy. Viruses 2018, 10, 501. [Google Scholar] [CrossRef] [PubMed]
- Koo, C.; Kaur, S.; Teh, Z.-Y.; Xu, H.; Nasir, A.; Lai, Y.-L.; Khan, E.; Ng, L.-C.; Hapuarachchi, H.C. Genetic Variability in Probe Binding Regions Explains False Negative Results of a Molecular Assay for the Detection of Dengue Virus. Vector Borne Zoonotic Dis. 2016, 16, 489–495. [Google Scholar] [CrossRef]
- Low, S.L.; Leo, Y.S.; Lai, Y.L.; Lam, S.; Tan, H.H.; Wong, J.C.C.; Tan, L.K.; Ng, L.C. Evaluation of Eight Commercial Zika Virus IgM and IgG Serology Assays for Diagnostics and Research. PLoS ONE 2021, 16, e0244601. [Google Scholar] [CrossRef]
- Kasbergen, L.M.R.; Nieuwenhuijse, D.F.; De Bruin, E.; Sikkema, R.S.; Koopmans, M.P.G. The Increasing Complexity of Arbovirus Serology: An in-Depth Systematic Review on Cross-Reactivity. PLoS Neglected Trop. Dis. 2023, 17, e0011651. [Google Scholar] [CrossRef]
- Musso, D.; Desprès, P. Serological Diagnosis of Flavivirus-Associated Human Infections. Diagnostics 2020, 10, 302. [Google Scholar] [CrossRef]
- Fortuna, C.; Remoli, M.E.; Rizzo, C.; Benedetti, E.; Fiorentini, C.; Bella, A.; Argentini, C.; Farchi, F.; Castilletti, C.; Capobianchi, M.R.; et al. Imported Arboviral Infections in Italy, July 2014–October 2015: A National Reference Laboratory Report. BMC Infect. Dis. 2017, 17, 216. [Google Scholar] [CrossRef]
- Baronti, C.; Piorkowski, G.; Charrel, R.N.; Boubis, L.; Leparc-Goffart, I.; de Lamballerie, X. Complete Coding Sequence of Zika Virus from a French Polynesia Outbreak in 2013. Genome Announc. 2014, 2, e00500-14. [Google Scholar] [CrossRef] [PubMed]
- Ministero della Salute. Direzione Generale della Prevenzione sanitaria. In Piano Nazionale della Prevenzione 2025; Ministero della Salute: Rome, Italy, 2020. [Google Scholar]
- Amraoui, F.; Failloux, A.-B. Chikungunya: An Unexpected Emergence in Europe. Curr. Opin. Virol. 2016, 21, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Rico-Mendoza, A.; Porras-Ramírez, A.; Chang, A.; Encinales, L.; Lynch, R. Co-Circulation of Dengue, Chikungunya, and Zika Viruses in Colombia from 2008 to 2018. Rev. Panam. Salud Pública 2019, 43, e49. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Morales, A.J.; Villamil-Gómez, W.E.; Franco-Paredes, C. The Arboviral Burden of Disease Caused by Co-Circulation and Co-Infection of Dengue, Chikungunya and Zika in the Americas. Travel Med. Infect. Dis. 2016, 14, 177–179. [Google Scholar] [CrossRef]
- Zammarchi, L.; Tappe, D.; Fortuna, C.; Remoli, M.E.; Günther, S.; Venturi, G.; Bartoloni, A.; Schmidt-Chanasit, J. Zika Virus Infection in a Traveller Returning to Europe from Brazil, March 2015. Eurosurveillance 2015, 20, 21153. [Google Scholar] [CrossRef]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef]
- Del Manso, M.; Caporali, M.; Bella, A.; Venturi, G.; Di Luca, M.; Giannitelli, S.; Ferraro, F.; Pezzotti, P.; Riccardo, F. Arbovirosi in Italia—Dengue N. 1—Novembre 2022; Istituto Superiore di Sanità: Rome, Italy, 2022. [Google Scholar]
- Drosten, C.; Göttig, S.; Schilling, S.; Asper, M.; Panning, M.; Schmitz, H.; Günther, S. Rapid Detection and Quantification of RNA of Ebola and Marburg Viruses, Lassa Virus, Crimean-Congo Hemorrhagic Fever Virus, Rift Valley Fever Virus, Dengue Virus, and Yellow Fever Virus by Real-Time Reverse Transcription-PCR. J. Clin. Microbiol. 2002, 40, 2323–2330. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and Serologic Properties of Zika Virus Associated with an Epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef]
| NRLA Cases Classification for DENV | ||||||
|---|---|---|---|---|---|---|
| Year | Confirmed | Probable | Excluded | NC | PRO | Total |
| Nov-2015 | 9 1 | 0 | 9 | 1 | 2 | 21 |
| 2016 | 46 1 | 5 | 96 | 19 | 11 | 177 |
| 2017 | 29 | 1 | 46 | 9 | 7 | 92 |
| 2018 | 17 1 | 0 | 57 | 7 | 3 | 84 |
| 2019 | 38 1 | 1 | 60 | 10 | 10 | 119 |
| 2020 | 0 | 0 | 10 | 0 | 1 | 11 |
| 2021 | 0 | 0 | 6 | 0 | 0 | 6 |
| Nov-2022 | 8 | 0 | 4 | 1 | 1 | 14 |
| Total | 146 | 7 | 288 | 48 | 35 | 524 |
| NRLA Cases Classification for ZIKV | ||||||
|---|---|---|---|---|---|---|
| Year | Confirmed | Probable | Excluded | NC | PRO | Total |
| Nov-2015 | 0 | NA | 14 | 2 | 2 | 18 |
| 2016 | 32 | NA | 212 | 17 | 11 | 272 |
| 2017 | 8 1 | NA | 68 | 8 | 7 | 91 |
| 2018 | 1 | NA | 72 | 9 | 3 | 85 |
| 2019 | 3 | NA | 94 | 9 | 10 | 116 |
| 2020 | 0 | NA | 10 | 1 | 0 | 11 |
| 2021 | 0 | NA | 11 | 0 | 0 | 11 |
| Nov-2022 | 0 | NA | 11 | 3 | 1 | 15 |
| Total | 44 | NA | 492 | 49 | 34 | 619 |
| DENV | Tot a Patients (N = 524) | Confirmed (N = 146) | Probable (N = 7) | Excluded (N = 288) | NC (N = 48) | PRO (N = 35) |
|---|---|---|---|---|---|---|
| Molecular test pos b/tested (%) | 36/318 (11.3%) | 36/95 (37.9%) | 0/4 (0.0%) | 0/173 (0.0%) | 0/26 (0.0%) | 0/20 (0.0%) |
| NS1 antigen ELISA pos/tested (%) | 78/311 (25.1%) | 76/110 (69.1%) | 0/5 (0.0%) | 2/124 (1.6%) | 0/44 (0.0%) | 0/28 (0.0%) |
| ELISA IgM system 1 pos/tested (%) | 38/121 (31.4%) | 26/29 (89.7%) | 2/3 (66.7%) | 4/67 (6.0%) | 1/12 (8.3%) | 5/10 (50.0%) |
| ELISA IgM system 2 pos/tested (%) | 27/65 (41.5%) | 11/14 (78.6%) | 1/2 (50.0%) | 7/37 (18.9%) | 4/7 (57.1%) | 4/5 (80.0%) |
| ELISA IgM system 3 pos/tested (%) | 103/358 (28.8%) | 79/101 (78.2%) | 3/5 (60.0%) | 6/181 (3.3%) | 5/43 (11.6%) | 10/28 (35.7%) |
| PRNT pos + bl c/tested (%) | 168 + 59/508 (33.1%) | 99 + 30/145 (68.3%) | 0 + 7/7 (0.0%) | 1 + 9/273 (0.4%) | 37 + 9/48 (77.1%) | 31 + 4/35 (88.6%) |
| ZIKV | Tot a Patients (N = 619) | Confirmed (N = 44) | Probable (N = 0) | Excluded (N = 492) | NC (N = 49) | PRO (N = 34) |
|---|---|---|---|---|---|---|
| Molecular test pos b/tested | 13/411 (3.1%) | 13/22 (59.1%) | NA | 0/345 (0.0%) | 0/25 (0.0%) | 0/19 (0.0%) |
| ELISA IgM system 4 pos/tested | 5/30 (16.7%) | 4/7 (57.1%) | NA | 0/11 (0.0%) | 0/9 (0.0%) | 1/3 (33.3%) |
| ELISA IgM system 5 pos/tested | 104/487 (21.4%) | 34/44 (77.3%) | NA | 41/363 (11.3%) | 12/48 (25.0%) | 17/32 (53.1%) |
| PRNT pos + bl c/tested | 95 + 54/594 (16%) | 38 + 5/44 (86%) | NA | 1 + 28/466 (0.2%) | 29 + 14/49 (59%) | 27 + 7/34 (79%) |
| Anti-ZIKV IgM ELISA System 5 | Anti-ZIKV PRNT | ||||||
|---|---|---|---|---|---|---|---|
| DENV confirmed (n = 98) | pos a | tested c | % | pos | bl b | tested | % |
| 10 | 106 | 9.4 | 0 | 31 | 108 | 28.7 | |
| Anti-DENV IgM ELISA System 1 | Anti-DENV IgM ELISA System 2 | Anti-DENV IgM ELISA System 3 | Anti-DENV PRNT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZIKV confirmed (n = 30) | pos a | tested c | % | pos | tested | % | pos | tested | % | pos | bl b | tested | % |
| 3 | 12 | 25.0 | 9 | 29 | 45.0 | 7 | 29 | 24.1 | 0 | 6 | 39 | 15.4 | |
| DENV | ≤3 Days | >3 to 7 Days | >7 Days | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pos | b.l. | Total | % | pos | b.l. | Total | % | pos | b.l. | Total | % | |
| Molecular | 11 | NA | 14 | 78.6 | 15 | NA | 37 | 40.5 | 14 | NA | 36 | 38.9 |
| NS1 ELISA | 3 | 0 | 5 | 60.0 | 30 | 1 | 35 | 88.6 | 22 | 4 | 39 | 66.7 |
| IgM ELISA | 6 | 0 | 11 | 54.5 | 31 | 6 | 44 | 84.1 | 41 | 2 | 49 | 87.8 |
| PRNT | 2 | 2 | 15 | 26.7 | 20 | 15 | 44 | 79.5 | 38 | 12 | 52 | 96.2 |
| ZIKV | ≤3 Days | >3 to ≤7 Days | >7 Days | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pos | b.l. | Total | % | pos | b.l. | Total | % | pos | b.l. | Total | % | |
| Molecular | 3 | NA | 4 | 75.0 | 15 | NA | 23 | 65.2 | 3 | NA | 19 | 15.8 |
| IgM ELISA | 1 | 0 | 3 | 33.3 | 9 | 3 | 14 | 85.7 | 16 | 0 | 16 | 100.0 |
| PRNT | 1 | 2 | 3 | 100.0 | 10 | 2 | 14 | 85.7 | 13 | 1 | 14 | 100.0 |
| DENV | Confirmed (n = 29) | Excluded (n = 66) | Total | |
|---|---|---|---|---|
| IgM ELISA system 1 | Positive | 30 | 4 | 34 |
| Negative | 6 | 67 | 73 | |
| Total | 36 | 71 | 107 | |
| Value (95% CI) | ||||
| Sensitivity | 83.33% (76.27, 90.39) | |||
| Specificity | 94.37% (90.00, 98.74) | |||
| PPV | 88.24% (82.13, 94.34) | |||
| NPV | 91.78% (86.58, 96.98) | |||
| Confirmed (n = 13) | Excluded (n = 37) | Total | ||
| IgM ELISA system 2 | Positive | 11 | 9 | 20 |
| Negative | 2 | 34 | 36 | |
| Total | 13 | 43 | 56 | |
| Value (95% CI) | ||||
| Sensitivity | 84.62% (75.17, 94.07) | |||
| Specificity | 79.07% (68.41, 89.72) | |||
| PPV | 55.00% (41.97, 68.03) | |||
| NPV | 94.44% (88.45, 100.44) | |||
| DENV | Confirmed (n = 85) | Excluded (n = 156) | Total | |
| IgM ELISA system 3 | Positive | 86 | 9 | 95 |
| Negative | 11 | 181 | 192 | |
| Total | 97 | 190 | 287 | |
| Value (95% CI) | ||||
| Sensitivity | 88.66% (84.99, 92.33) | |||
| Specificity | 95.26% (92.81, 97.71) | |||
| PPV | 90.53% (87.15, 93.90) | |||
| NPV | 94.27% (91.58, 96.96) | |||
| Notes: 9 of DENV confirmed cases showed a positive molecular but negative IgM ELISA results (4 for system 1, 1 for system 2 and 4 for system 4). | ||||
| ZIKV | Confirmed (n = 37) | Excluded (n = 344) | Total | |
| IgM ELISA system 5 | Positive | 44 | 44 | 88 |
| Negative | 10 | 346 | 356 | |
| Total | 54 | 390 | 448 | |
| Value (95% CI) | ||||
| Sensitivity | 81.48% (77.87, 85.09) | |||
| Specificity | 88.72% (85.78, 91.65) | |||
| PPV | 50.00% (45.35, 54.65) | |||
| NPV | 97.19% (95.65, 98.73) | |||
| Notes: 4 of ZIKV confirmed cases showed a positive molecular result, but negative IgM ELISA results for system 5. | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merakou, C.; Amendola, A.; Fortuna, C.; Marsili, G.; Fiorentini, C.; Argentini, C.; Benedetti, E.; Rezza, G.; Maraglino, F.; Del Manso, M.; et al. Diagnosis of Imported Dengue and Zika Virus Infections in Italy from November 2015 to November 2022: Laboratory Surveillance Data from a National Reference Laboratory. Viruses 2024, 16, 50. https://doi.org/10.3390/v16010050
Merakou C, Amendola A, Fortuna C, Marsili G, Fiorentini C, Argentini C, Benedetti E, Rezza G, Maraglino F, Del Manso M, et al. Diagnosis of Imported Dengue and Zika Virus Infections in Italy from November 2015 to November 2022: Laboratory Surveillance Data from a National Reference Laboratory. Viruses. 2024; 16(1):50. https://doi.org/10.3390/v16010050
Chicago/Turabian StyleMerakou, Christina, Antonello Amendola, Claudia Fortuna, Giulia Marsili, Cristiano Fiorentini, Claudio Argentini, Eleonora Benedetti, Gianni Rezza, Francesco Maraglino, Martina Del Manso, and et al. 2024. "Diagnosis of Imported Dengue and Zika Virus Infections in Italy from November 2015 to November 2022: Laboratory Surveillance Data from a National Reference Laboratory" Viruses 16, no. 1: 50. https://doi.org/10.3390/v16010050
APA StyleMerakou, C., Amendola, A., Fortuna, C., Marsili, G., Fiorentini, C., Argentini, C., Benedetti, E., Rezza, G., Maraglino, F., Del Manso, M., Bella, A., Pezzotti, P., Riccardo, F., Palamara, A. T., Venturi, G., & The Arbovirus Working Group. (2024). Diagnosis of Imported Dengue and Zika Virus Infections in Italy from November 2015 to November 2022: Laboratory Surveillance Data from a National Reference Laboratory. Viruses, 16(1), 50. https://doi.org/10.3390/v16010050






